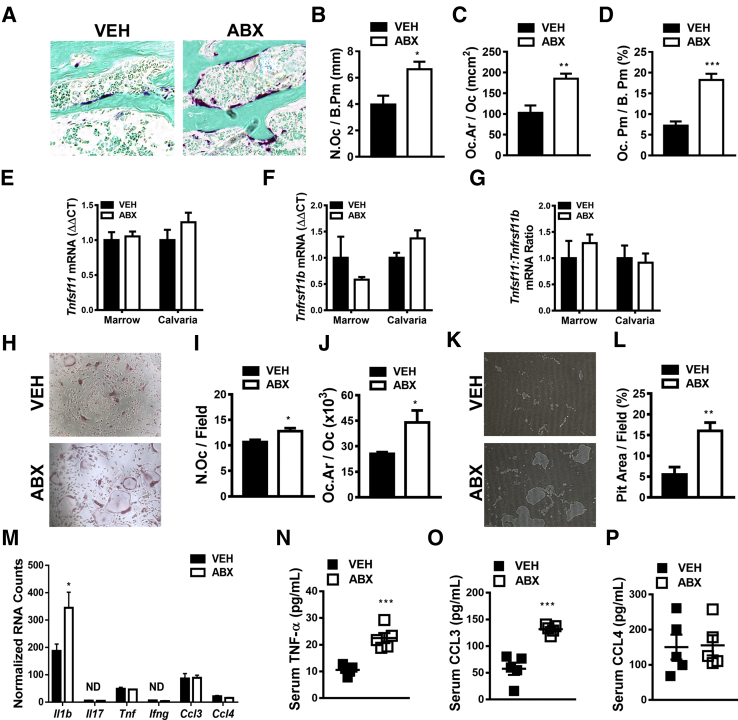
Figure 5.
Antibiotic (ABX) perturbation of gut microbiota effects on osteoclastogenesis and local/systemic proinflammatory cytokines. Twelve-week–old male vehicle (VEH)– and ABX-treated mice were euthanized; specimens were harvested for analysis. A–D: Histomorphometric analyses of osteoclast cellular end points were performed in the trabecular bone secondary spongiosa of tartrate-resistant acid phosphatase (TRAP)–stained proximal tibia sections; TRAP+ cell lining bone with three or more nuclei designated an osteoclast. A: Representative images of TRAP-stained (fast green counterstain) secondary spongiosa. B: Osteoclast number per bone perimeter (N.Oc/B.Pm). C: Osteoclast area/size (Oc.Ar/Oc). D: Osteoclast perimeter per bone perimeter (Oc.Pm/B.Pm). E–G: Real-time quantitative RT-PCR analysis in bone marrow and calvaria to assess alterations in the RANKL/OPG axis: Tnfsf11(Rankl) mRNA (E), Tnfrsf11b(Opg) mRNA (F), and Tnfsf11(Rankl):Tnfrsf11b(Opg) ratio (G). Relative quantification of mRNA was performed via the comparative CT method (ΔΔCT); Gapdh was used as an internal control. Data are expressed as fold difference relative to VEH. H–J: Hematopoietic cells were isolated from marrow, and CD11b sorting was performed to enrich for CD11bneg OCP cells. CD11bneg OCP cells were stimulated with 25 ng/mL colony stimulating factor 1 (CSF1) and 50 ng/mL RANKL to induce osteoclastogenesis. Cytomorphometric cellular end points were analyzed in TRAP-stained cultures. TRAP+ cells with three or more nuclei were considered osteoclasts. H: Representative images of OCP cultures stimulated with CSF1 and RANKL media. I: Number of osteoclasts per field of view (N.Oc/Field). J: Average osteoclast size (Oc.Ar/Oc). K and L: Hematopoietic cells were isolated from marrow, and CD11b sorting was performed to enrich for CD11bneg OCP cells. CD11bneg OCP cells were stimulated with 25 ng/mL CSF1 and 50 ng/mL RANKL to induce osteoclastogenesis. Osteoclast resorption activity was assessed by quantifying pit area in Osteo Assay plates. K: Representative images of pit area. L: Pit area per field area (percentage). M: NanoString analysis was performed to assess proinflammatory cytokines in femur bone marrow. Data were normalized to the geometric means of spiked-in positive controls and internal control genes. Absolute quantification of RNA expressed as normalized RNA counts. N–P: Serum was isolated from whole blood; enzyme-linked immunosorbent assay analysis of TNF-α (N), CCL3 (O), and CCL4 (P). Unpaired t-test was used. Data are expressed as means ± SEM. n = 4 per group (A–D and H–M); n = 4 to 5 per group (E–G); n = 5 per group (N–P). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus vehicle. Original magnification, ×200 (A, H, and K). ND, not detectable.