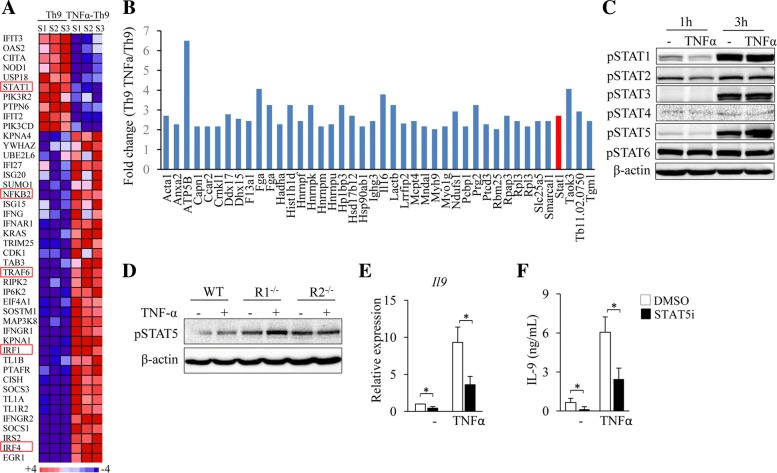
Fig. 5.
TNF-α enhances Th9 cell differentiation through STAT5. (a) The RNA-seq data mentioned in Fig. 2a were used. Pink-blue heatmap shows the log2-fold change of the differentially expressed genes of signaling pathways that are potentially involved in TNF-α-induced Th9 cell differentiation. (b) Naïve CD4+ T cells were cultured under Th9 polarizing conditions with or without addition of TNF-α for 3 h. Immunoprecipitation (IP) was performed by using anti-TNFR2 and proteins in the immune precipitate were analyzed by mass spectrometry (MS). Shown are the fold changes (TNF-α-Th9/Th9) of proteins with 2-fold cut-off. (c) Naïve CD4+ T cells were cultured under Th9 polarizing conditions with or without addition of TNF-α for 1 or 3 h. Western-blots examined the protein levels of the phosphorylated STAT family members (pSTAT1–6). β-actin was used as a loading control. (d) Naïve CD4+ T cells from WT, R1−/− or R2−/− mice were cultured under Th9 polarizing conditions with or without the addition of TNF-α for 3 h. Western-blots analyzed pSTAT5 and β-actin in T cells. (e, f) Naïve CD4+ T cells were cultured under Th9 polarizing conditions in the presence or absence of TNF-α with or without (DMSO) addition of STAT5 inhibitor (STAT5i) for 3 days. IL-9 expression was examined by qPCR (e) and ELISA (f). Data are representative of three (c, d) independent experiments or presented as mean ± SD of three (e, f) independent experiments. *P < 0.05