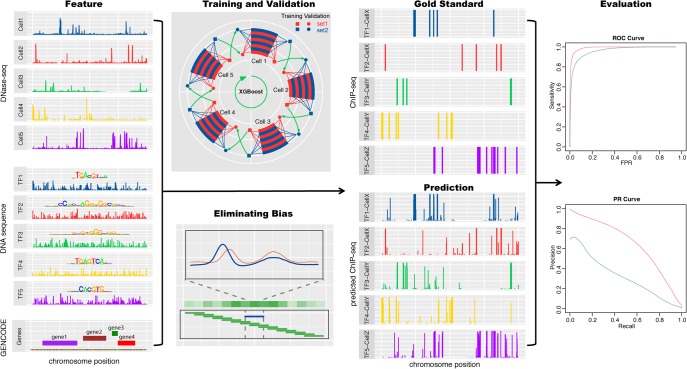
Figure 1.
Overview of the TF binding profile prediction. The DNase-seq signals, TF binding motifs, and distances to the nearest genes are the input features. After eliminating sequencing and cell-type–specific DNase-seq biases, XGBoost models are trained and validated in a cross-cell type and cross-chromosome fashion to exploit the limited data and avoid overfitting. In the training and validation panel, two randomly partitioned chromosome sets are shown in red and blue. The square sets are used to train XGBoost models, and the validation circle sets are used to select hyperparameters. The evaluation is based on the AUROC and AUPRC between the genome-wide predictions and the gold standard ChIP-seq profiles.