Fig. 1. A genome-wide saturation mutagenesis screen for Plasmodium falciparum.
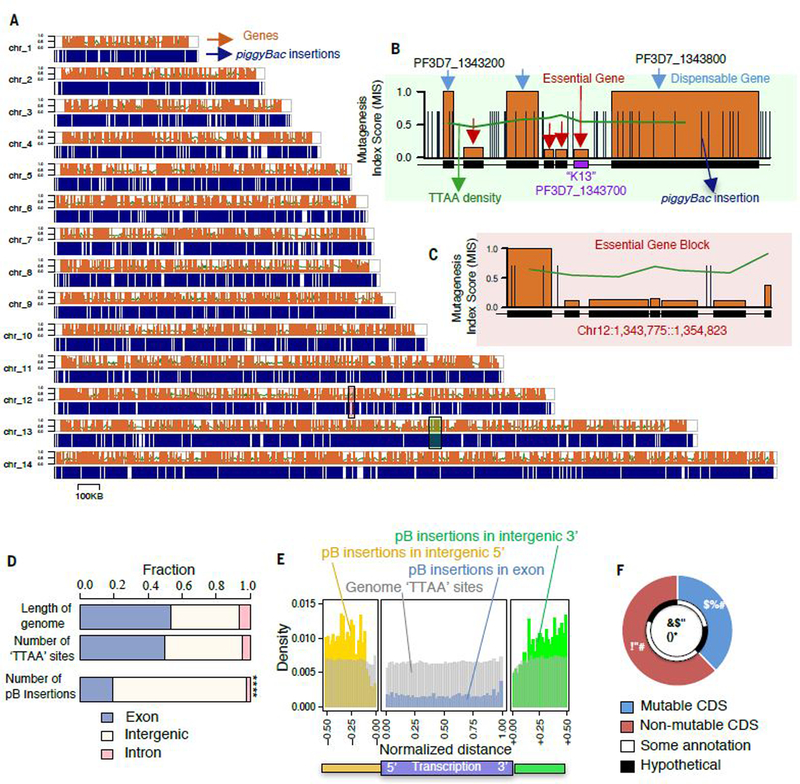
(A) Chromosomal map displays 38,173 piggyBac insertion sites from all mutants evenly distributed throughout the genome. (B) High-resolution map of a ~50 KB region of chromosome 13 depicts an essential gene cluster, including K13, flanked by dispensable genes with multiple CDS-disrupting insertions. (C) High-resolution map of a ~20 KB region without insertions includes three conserved genes of unknown function (PF3D7_1232700, PF3D7_1232800, PF3D7_1232900) and a putative nucleotidyltransferase (PF3D7_1232600) (see also fig. S5). (D) A plot of all piggyBac insertions revealed significantly fewer insertions were recovered from exon-intron regions compared to the proportion of available TTAA sites (see also fig. S1D) (p < 2.2e-16, Fisher’s Exact test). (E) Density of piggyBac insertion-site distribution revealed threefold fewer insertions recovered in transcriptional regions (blue) than intergenic 5ʹ (yellow) and 3ʹ (green) regions, depicted as relative distance upstream and downstream to a gene, respectively. (F) This study determined that under ideal culture conditions for asexual blood-stage growth, 38% of genes in the P. falciparum genome have mutable CDSs, while 62% of genes have non-mutable CDSs.
