Fig. 4. Chromosomal syntenic breakpoints are enriched in dispensable genes.
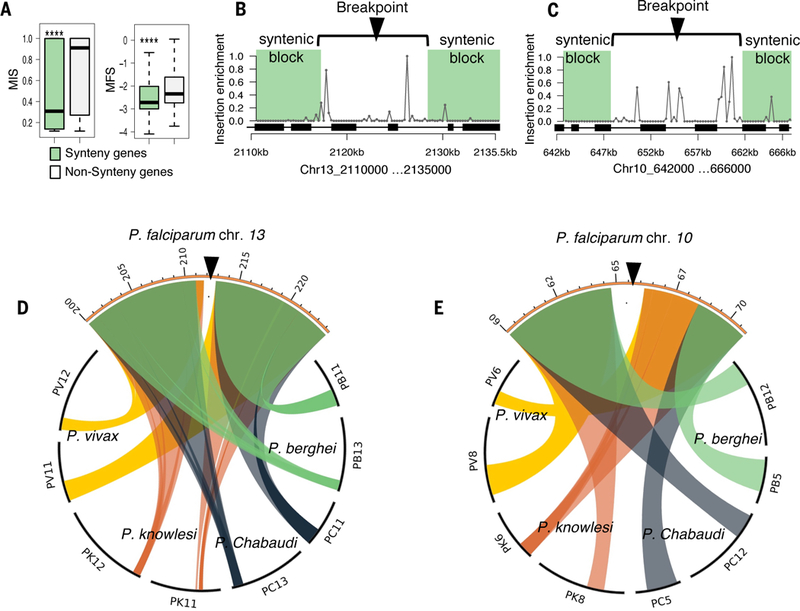
(A) Genes within conserved syntenic blocks have significantly lower MIS and MFS (Wilcoxon p < 2.2e-16). Syntenic genes or “syntenic block” is defined as at least three genes in the same order on the same chromosome as their orthologs in another species within a 25-kb search window. (B and C) Scatter plots show the insertion site enrichment along two syntenic breakpoints (Ch13:2,110,000 −2,135,000, Chr10: 642000–666000). Each gap in synteny (white area) is enriched for piggyBac insertions while flanked by essential regions (green shading); black boxes represent the location of CDS. (D and E) Circos plots indicate the syntenic blocks of P. falciparum in relation to other Plasmodium spp. (P. berghei, P. chabaudi, P. knowlesi, P. vivax).
