Summary
Successive cell divisions during embryonic cleavage create increasingly smaller cells, so intracellular structures must adapt accordingly. Mitotic spindle size correlates with cell size, but the mechanisms for this scaling remain unclear. Using live cell imaging, we analyzed spindle scaling during embryo cleavage in the nematode Caenorhabditis elegans and sea urchin Paracentrotus lividus. We reveal a common scaling mechanism, where the growth rate of spindle microtubules scales with cell volume, which explains spindle shortening. Spindle assembly timing is however constant throughout successive divisions. Analyses in silico suggest that controlling the microtubule growth rate is sufficient to scale spindle length and maintain a constant assembly timing. We tested our in silico predictions to demonstrate that modulating cell volume or microtubule growth rate in vivo induces a proportional spindle size change. Our results suggest that scalability of the microtubule growth rate when cell size varies adapts spindle length to cell volume.
Introduction
Eukaryotic cells range in size over six orders of magnitude. Regardless of size from the smallest unicellular eukaryote Ostreococcus tauri (<1 μm) to long motor neurons (>1 m long), all cells contain a common set of organelles and subcellular structures. This requirement mandates a strict scaling between these intracellular components and cell size. One striking example of this scaling is the progressive decrease in cell volume that occurs during cleavage of early embryos. Early embryonic development usually occurs without an increase in the overall embryo volume. Each round of cell division halves the size of blastomeres, so blastomere volume can decrease by three orders of magnitude from single celled-zygotes to multicellular gastrulae (O’Farrell et al., 2004). Thus, intracellular structures and organelles must adapt to this wide range of cell dimensions by adjusting their spatial distribution and/or size to remain functional (West and Brown, 2005). Organelles known to exhibit size adaptation to cell volume variations include nuclei, cilia, endoplasmic reticulum, and cytoskeletal assemblies like the metaphase spindle (reviewed in Chan and Marshall, 2010, 2012; Levy and Heald, 2012; Marshall, 2015b; Reber and Goehring, 2015).
The metaphase spindle is a bipolar structure composed of highly dynamic microtubules. Adapting spindle length to cell volume enables accurate chromosome segregation and spindle orientation to accommodate different scales (Dumont and Mitchison, 2009a; Holmfeldt et al., 2006; Lancaster et al., 2013). Spindle length scaling among different organisms can occur through biochemical variations in cytoplasmic content. For example, a single amino-acid change in the microtubule-severing protein katanin primarily accounts for the spindle length difference observed between oocytes from Xenopus laevis and the smaller closely related frog Xenopus tropicalis (Brown et al., 2007; Loughlin et al., 2011). In contrast, the biochemical composition of different sized blastomeres from a given species is assumed to be constant (Mitchison et al., 2015). During cleavage of the large X. laevis embryo, spindle length remains constant for the first five divisions and then decreases linearly with blastomere radius for the next 5–7 divisions (Wuhr et al., 2008). In contrast, the smaller C. elegans embryo shows spindle length proportional to cell length from the first division throughout cleavage (Decker et al., 2011; Hara and Kimura, 2009, 2013). Seminal experiments using artificially encapsulated extracts from X. laevis oocytes or embryos demonstrated that spindle length directly corresponds to the size of the encapsulating droplet (Good et al., 2013; Hazel et al., 2013). These experiments accurately recapitulated the spindle scaling observed in intact embryos with a linear relationship between spindle length and droplet radius in small droplets and an upper limit to spindle length in large droplets. Intrinsic spindle mechanisms, such as balancing force between opposed motors, may account for the upper limit of spindle length scaling (Dumont and Mitchison, 2009a, b; Reber and Goehring, 2015). In contrast, spindle extrinsic mechanisms, such as component limitation, have been proposed to explain how different cytoplasm volumes with a given composition may produce different spindle lengths (Goehring and Hyman, 2012; Marshall, 2015a; Mitchison et al., 2015; Reber and Goehring, 2015; Reber and Hyman, 2015). In C. elegans early embryos, decreasing spindle length correlates with a progressive reduction in the amount of centrosomal components and with a decaying gradient of the microtubule-associated protein TPXL-1 (ortholog of TPX2) along spindle microtubules (Greenan et al., 2010). Experiments performed in Xenopus egg or embryo extracts suggest that titration of tubulin heterodimers, limited amount/activity of the microtubule polymerase XMAP215, or cortical sequestration of Importin α, the negative regulator of the microtubule depolymerase KIF2A, could participate in spindle length scaling (Reber et al., 2013; Wilbur and Heald, 2013). Yet, it remains unclear how the limited supply of these factors controls spindle size.
The limited supply or local inactivation of key factor(s) could directly regulate spindle microtubule dynamics to efficiently control spindle length (Andersen, 2000; Andersen et al., 1997; Desai and Mitchison, 1997; Goshima et al., 2005b; Picone et al., 2010; Tournebize et al., 2000; Verde et al., 1992). Both in vitro and in vivo, microtubules alternate between phases of growth and shrinkage in a process called dynamic instability (Mitchison and Kirschner, 1984). Four parameters define these dynamic properties of microtubules, the velocities of growth and shrinkage and the frequencies of transitions between these two states called catastrophe and rescue (Kirschner and Mitchison, 1986; Walker et al., 1988). Typically, long interphasic microtubules display low catastrophe and high rescue frequencies (Belmont et al., 1990; Verde et al., 1992; Verde et al., 1990). Microtubules in this state are “unbounded” and continuously elongate over time. In contrast, microtubules shorten upon mitotic entry, due to a sharp increase in catastrophe frequency and a significant decrease in rescue events. In this “bounded” regime, microtubules tend to disassemble, and their average growth velocity, termed J, is negative. A steady population of microtubules can only be maintained because a finite fraction is continuously nucleated de novo (Mitchison et al., 2015; Verde et al., 1992). This regime establishes a distribution of microtubule lengths to dictate a steady state spindle size. Therefore, precise control of microtubule dynamics during mitosis in cleaving embryos becomes an attractive candidate to adjust spindle length for blastomere size. However, the functional link between microtubule dynamics and spindle length scaling as a function of cell volume during embryo cleavage remains unknown.
Results
Microtubule Dynamics are Modulated During C. elegans Embryo Cleavage
We first determined the potential relationship among metaphase spindle length, cell volume, and microtubule dynamics from the 1- to the 16-cell stage in cleaving C. elegans embryos. We combined high-temporal single plane confocal microscopy and 2-photon 3D-volumetric reconstructions of live embryos expressing GFP-tagged microtubules or a plasma membrane marker respectively (Figure 1A and S1A,B). In line with previous studies, we found that spindle length and cell volume progressively decreased in a sub-proportional manner across early embryogenesis in C. elegans (Figure S1C–F) (Decker et al., 2011; Greenan et al., 2010; Hara and Kimura, 2009, 2013). To determine if microtubule dynamics vary concomitantly with spindle length and cell volume, we generated kymographs of individual microtubules from our high frame-rate, single-plane confocal series of embryos expressing GFP-tagged β-tubulin (Figure 1B). This approach allowed us to identify phases of elongation and shortening and to measure the rate and duration of individual phases for all analyzed microtubules. We observed that microtubule growth rates and durations were significantly higher for astral microtubules than spindle microtubules in all blastomeres from the 1- to the 16-cell stage, in agreement with published results for the one-cell stage (Figure 1C) (Srayko et al., 2005). In addition, we found that microtubule dynamics’ parameters for both types of microtubules varied throughout development. While we found a notable but mild decrease in shrinkage duration between the 1- and the 16-cell stage, the most striking variation across divisions was the continuous and significant decrease in microtubule growth rate for both astral and spindle microtubules (Figure 1C). Importantly, we obtained similar rates and frequencies using a different promoter to drive expression of our GFP-tagged β-tubulin reporter or mCherry-tagged β-tubulin (Figure S2 and Table S1). These results suggest that microtubule dynamics vary throughout early embryonic divisions, with a distinct decrease in the microtubule growth rate associated with decreased blastomere volume.
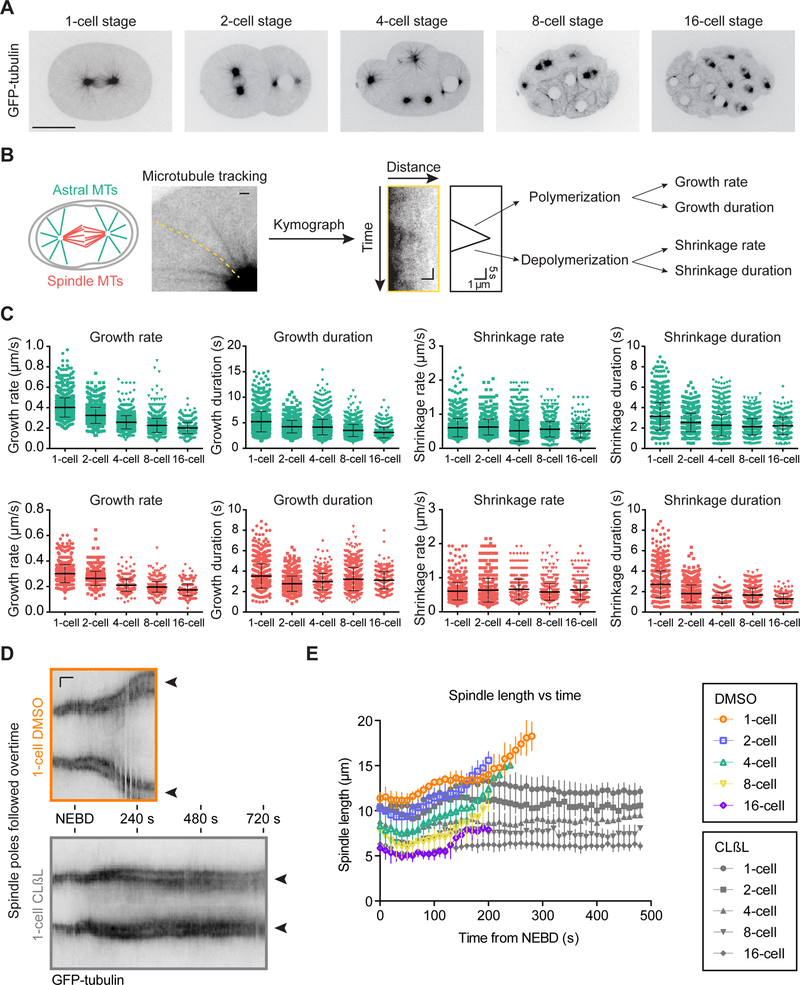
Figure 1. Astral and Spindle Microtubule Dynamics Vary During C. elegans Embryo Cleavage.
(A) Still frames from confocal live imaging of C. elegans embryos expressing GFP-tagged β-tubulin during the first five embryonic divisions (1- to 16-cell stage). Images correspond to a single focal plane. Scale bar, 20 μm.
(B) Schematic representation of parameter extraction for microtubule dynamics from individual astral (green) or spindle (red) microtubule kymographs. One representative example of a kymograph shown of an individual astral microtubule tracked over time. Horizontal scale bars, 1 μm, vertical scale bar (kymograph), 5 s.
(C) Microtubule dynamics parameters for astral (green) and spindle (red) microtubules plotted at each cleavage stage (1- to 16-cell stage). Each dot corresponds to an individual microtubule. For both microtubule populations, each parameter significantly varies between stages (one-way ANOVA: p<0.01). Only microtubule growth rates of both microtubule populations exhibit significant and continuous decrease at each stage (one-way ANOVA with Tukey’s multiple comparison tests: p<0.01). All parameters including sample size, mean, and SD for the four astral and spindle microtubule dynamics parameters at the different stages are listed in Table S1. (Horizontal bars, mean; error bars, SD; n(microtubules)≥288 per cleavage stage; n (embryos)≥12).
(D) Kymographs extracted from confocal live imaging of C. elegans embryos expressing GFP-tagged β-tubulin during the first embryonic division (one-cell stage) treated with DMSO (control, top) or with 20 μM of the proteasome inhibitor Clasto-Lactacystin β-Lactone (CLβL diluted in DMSO, bottom) to inhibit the metaphase-to-anaphase transition. Spindles poles (black arrowheads) are visible as two dark stripes that separate during anaphase in controls and stay at the same distance over time in CLβL-treated embryos. Timings relative to NEBD monitored by diffusion of free GFP-tagged β-tubulin in the nuclear area. Horizontal scale bar, 50s; Vertical scale bar, 5 μm.(E) Spindle length (aster-to-aster distance) plotted over time in DMSO (colors) and CLβL (grey) treated embryos at each cleavage stage (1- to 16-cell stage). Timings relative to NEBD. (n≥5 spindles per stage in each condition; error bars, SD).
To investigate a potential link between these variations in microtubule dynamics and spindle length, we first determined if the length of metaphase spindles reached a steady state or if they were limited by the duration of the cell cycle and the time available before anaphase onset. Indeed, cell divisions in cleaving embryos of most species are particularly rapid, and spindle length increases continuously from nuclear envelope breakdown (NEBD) to anaphase onset (Mitchison et al., 2015). However, delaying anaphase onset in C. elegans embryos using a proteasome inhibitor did not induce spindle elongation above the normal metaphase spindle length at any analyzed stage (Figure 1D,E) (Labbe et al., 2004). Moreover, the measured microtubule dynamics parameters indicate that all microtubules are in the “bounded” regime (J<0) at every stage of embryo cleavage, which is compatible with a steady state (Figure S2B and Table S1A,C) (Verde et al., 1992). These results suggest that mitotic spindles in early C. elegans embryos reach a steady state length before anaphase onset that is independent of the duration of mitosis. Overall, our results demonstrate that microtubule dynamic instability evolves during embryo cleavage with the microtubule growth rate continuously decreasing in parallel with the progressive reduction of steady state spindle length.
Spindle Microtubule Growth Rate Adapts to Cell Volume in C. elegans
We next asked if variations in microtubule dynamics parameters during early embryogenesis were either a direct consequence of cell size changes or under the control of a developmental program. Therefore we analyzed the direct correlation between cell volume and microtubule dynamics parameters (Figure 2A,B). This analysis revealed that the growth rate of spindle microtubules most significantly correlated with cell volume (Figure 2B). So, we tested if the decrease in cell volume of dividing blastomeres solely accounted for this relationship or if cell fate patterning and developmental timing also contributed. Since C. elegans zygotes divide asymmetrically, patterning is already established at the 2-cell stage by asymmetric distribution of cell fate determinants (Nance, 2005; Rose and Kemphues, 1998). The first division of the C. elegans zygote produces a large anterior AB blastomere from which most somatic tissues emerge and a small posterior P1 blastomere from which the germline originates (Figure 2A) (Sulston et al., 1983). We used a fast acting temperature sensitive (ts) mutant of formin CYK-1 to induce cytokinesis failure in the zygote (Figure 2C) (Canman et al., 2008; Davies et al., 2014). In control 2-cell stage embryos, AB and P1 blastomeres display shorter spindles than the 1-celled zygote, which correlates with slower microtubule growth in both blastomeres (Figure 2D). At the restrictive temperature (25°C), cyk-1(ts) mutant zygotes exhibit cytokinesis failure and form abnormal one-cell embryos that kinetically correspond to 2-cell stage embryos but possess the cytoplasmic content and cell fate determinants of both AB and P1 blastomeres. These abnormal embryos assembled tetrapolar spindles with microtubules that grew at a rate indistinguishable from normal one-cell zygotes (Figure 2C,D). Thus cell fate determinants and developmental timing do not make a major contribution to set the spindle microtubule growth rate in cleaving C. elegans embryos. To further demonstrate the effect of cell volume on spindle microtubule growth rate, we generated abnormally large zygotes. We generated embryos depleted of the fucosyltransferase C27D9.1, which does not have any known direct effect on microtubules and leads to the formation of abnormally large embryos (Hara and Kimura, 2009; Sonnichsen et al., 2005). In these extra-large zygotes, we found longer spindles and a significantly higher growth rate of spindle microtubules than in control normal-sized zygotes (Figure 2C,D). Altogether, these results show that the observed progressive reduction in the growth rate of spindle microtubules is directly linked to cell volume during embryonic cleavage and independent of developmental timing or cell fate.
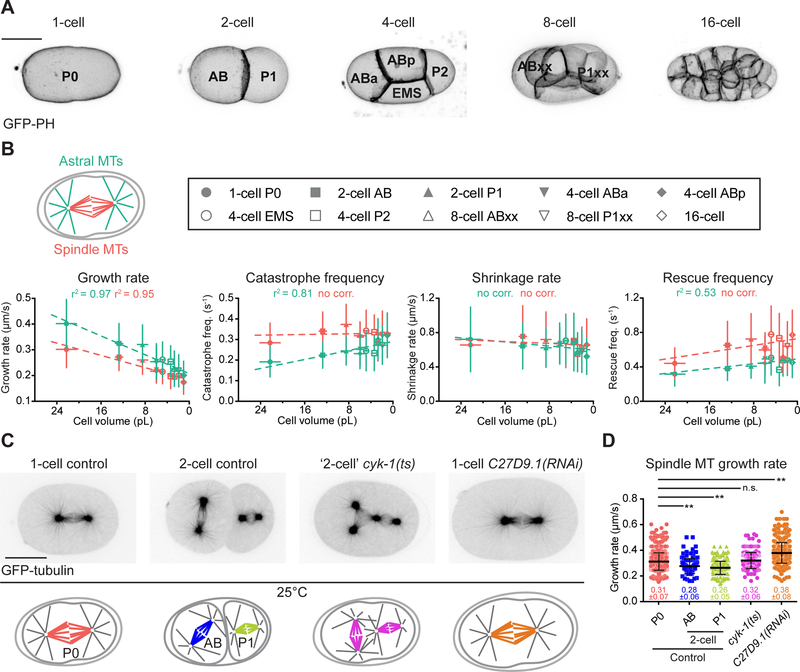
Figure 2. Astral and Spindle Microtubule Dynamics Vary with Cell Volume During C. elegans Embryo Cleavage.
(A) Still frames from live 2-photon imaging of C. elegans embryos expressing a GFP-tagged plasma membrane probe (Pleckstrin Homology (PH) domain) during the first five embryonic divisions (1- to 16-cell stage). Images correspond to maximal projections of z-stacks covering the entire thickness of the embryo. Scale bar, 20 μm. Blastomere names are indicated except for the 16-cell stage. At the 8-cell stage, progeny of AB and P1 were grouped together as ABxx and P1xx respectively.
(B) Mean astral (green) and spindle (red) microtubule dynamics parameters: mean microtubule growth rate, catastrophe frequency, shrinkage rate and rescue frequency (from Figure 1C) for each type of blastomere plotted over the corresponding average cell volumes (from Figure S1). Key for different blastomeres shown in the top box. Dotted lines correspond to the linear regression curves. Pearson correlation coefficient (r2) is indicated at the top of each graph if p ≤ 0.01 (no corr. is indicated otherwise).
(C) From left to right, still images from confocal live imaging of C. elegans control one-cell embryo, control 2-cell embryo, thermosensitive (ts) mutant embryo of the formin cyk-1 at the ‘2-cell’ stage after P0 cytokinesis failure and abnormally large C27D9.1(RNAi)-treated embryo. All express GFP-tagged β-tubulin. Corresponding schematics with color-coding for spindle microtubules in different conditions shown at the bottom. Scale bar, 20 μm.
(D) Spindle microtubule growth rates measured at 25°C (restrictive temperature for the cyk-1(ts) mutant) for the indicated conditions. Color-coding for the different conditions corresponds to the schematics in (C). (Error bars, SD; one-way ANOVA with Dunnett’s multiple comparisons test, **: p≤0.01, n.s.: p>0.05).
Spindle Length Scales with Microtubule Growth Rate in C. elegans
We then asked if variations between dynamics parameters of astral and spindle microtubules and cell volume could underlie spindle length scaling during early embryogenesis. Strikingly, we found that the growth rate of both astral and spindle microtubules was the only parameter exhibiting a distinct, continuous and linear relationship with spindle length throughout cleavage (Figure 3A–C). This observation suggests that modulating the growth rate of one or both microtubule populations could be a key determinant for spindle length adaptation in cleaving embryos. However, analysis of this scaling in C27D9.1-depleted extra-large zygotes revealed that only spindle microtubule growth rate increased with spindle length (Figure 3B), while astral microtubule growth rate remained similar to controls (Figure 3C and Figure S1G,H) (0.41±0.08 μm/s versus 0.40±0.09 μm/s, p=0.087 unpaired t-test). These results suggest that spindle, but not astral, microtubule dynamics determine spindle length in cleaving embryos.
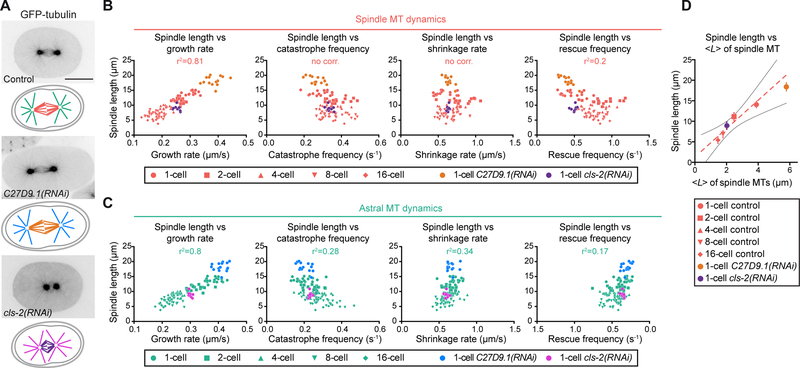
Figure 3. Spindle Length Scales with Spindle, but not Astral, Microtubule Growth Rate.
(A) From top to bottom (left to right), still images from confocal live imaging of C. elegans control one-cell embryo, abnormally large C27D9.1(RNAi)-treated embryo and cls-2(RNAi)-treated embryo. All expressing GFP-tagged β-tubulin. Corresponding schematics with color-coding for astral and spindle microtubules in the different conditions shown underneath each image. Scale bar, 20 μm.
(B) Spindle length plotted over spindle microtubule dynamics parameters in the indicated cleavage stage or condition. Key for different stages and conditions indicated at the bottom of the graphs. (Control: red, C27D9.1(RNAi): orange, cls-2(RNAi): purple). Each dot represents the spindle length represented over mean microtubule dynamics parameters measured in an individual blastomere. (n(cells)≥10 with n(events/cell)≥60). Pearson correlation coefficient (r2) for the control condition indicated at the top of each graph if p≤0.01 (no corr. is indicated otherwise).
(C) Same as (B) for astral microtubules. (Control: green, C27D9.1(RNAi): blue, cls-2(RNAi): magenta).
(D) Average theoretical microtubule length <L> plotted over experimentally measured average spindle length in the indicated conditions (Control: red, C27D9.1(RNAi): orange, cls-2(RNAi): purple). with Vg: Growth rate, Vs: Shrinkage rate, fc: Catastrophe frequency, fr: Rescue frequency. Dotted red line and black lines represent the linear regression and 95% confidence interval respectively.
As the growth rate of spindle microtubules decreased progressively with cell volume during embryo cleavage and concomitant with spindle length, we hypothesized that modulating this parameter is sufficient to scale spindle length to cell volume during embryo cleavage. We calculated the average length <L> predicted from microtubule dynamics parameters (Figure 3D, S2B,C and Table S1) (Verde et al., 1992). In control embryos, <L> scaled linearly with our experimentally measured spindle lengths from the 1- to the 16-cell stage. This scaling implies that spindle length is a multiple of <L> and that the multiplicative factor remains relatively constant as cell size evolves. Interestingly, the predicted average length <L> of spindle microtubules in C27D9.1-depleted embryos fitted well with the abnormally long spindles in these extra-large zygotes. Thus, spindle length varies proportionally to the overall microtubule dynamics in cleaving embryos. Since rescue frequency is low but catastrophe frequency and shrinkage rate are almost constant in cleaving C. elegans embryos, our results demonstrate that <L> is primarily determined by the microtubule growth rate (Figure 3B,D and Table S1). Our results support the idea that spindle microtubule growth rate is a key regulator to scale spindle length to cell volume.
To directly test this hypothesis, we experimentally reduced spindle microtubule growth rate in zygotes and measured spindle length. We analyzed zygotes partially depleted of the microtubule-associated protein (MAP) CLS-2 (ortholog of CLASP, Figure 3A–C) (Cheeseman et al., 2005; Espiritu et al., 2012), since CLS-2 promotes microtubule assembly in vitro and in vivo (Maton et al., 2015). Consistent with this, in partial cls-2(RNAi) conditions, spindle microtubules displayed a significantly reduced growth rate compared to in control zygotes (0.25±0.06 versus 0.3±0.07 μm/s, p<0.001 unpaired t-test) (Figure 3B and Table S1). Rescue frequency also slightly increased, while catastrophe frequency and shrinkage rate remained unaltered. As we predicted, abnormally short but functional spindles assembled under this condition. Strikingly, both the average theoretical length of microtubules <L> and spindle length in these depleted zygotes were concomitantly reduced to levels corresponding to that of 4-cell stage control blastomeres (Figure 3B–D). Taken together, our results show that microtubule dynamics parameters, specifically the growth rate of spindle microtubules, vary proportionally to cell volume and are key factors that control spindle length during C. elegans embryo cleavage.
Spindle Length and Microtubule Growth Rate Scale with Cell Volume in the Sea Urchin P. lividus
We next wondered whether the scaling relationship among cell volume, spindle length, and microtubule growth rate identified in C. elegans also takes place in other species. Sea urchin (Paracentrotus lividus) embryos are approximately 20 times larger in volume than C. elegans embryos. P. lividus zygotes are spherical and divide symmetrically and synchronously until the 8-cell stage (Figure 4A) (Agrell, 1956; Parisi et al., 1978). First, we microinjected P. lividus eggs with fluorescently labeled tubulin (ATTO 565-tubulin, Figure S3A–F), which then underwent fertilization. We examined the first six rounds of embryonic cleavages by live confocal microscopy (Movie S1 and S2). Consistent with previous reports for other echinoderms (Crowder et al., 2015), spindle length and cell volume progressively decreased across early embryogenesis in P. lividus (Figure S3G–H and Table S2). Spindle lengths measured in ATTO 565-tubulin injected live embryos were indistinguishable from those in control non-injected fixed embryos (Figure S3B,D). To determine if microtubule dynamics also varied concomitantly with spindle length and cell volume, we generated kymographs of individual microtubules to measure spindle microtubule dynamics parameters. In line with our results from C. elegans embryos, microtubule dynamics parameters varied during P. lividus cleavage, but the overall microtubule dynamics remained in the bounded regime at every stage (Figure 4B and Table S2). We observed a continuous and significant decrease in the growth rate of spindle microtubules across P. lividus embryo cleavage (Figure 4B). This decrease correlated with the progressive reduction in blastomere volume and spindle length (Figure 4C,D). We observed similar spindle lengths and growth rates using another fluorescent microtubule probe (the 3XmCherry-tagged ensconsin microtubule-binding domain, 3XmCherry-EMTB (Faire et al., 1999), Figure S3A–F). These results suggest that microtubule growth rate is a key determinant of spindle length in cleaving sea urchin embryos. Importantly, both spindle length and spindle microtubule growth rate were comparable in vegetal micromeres (which result from strong asymmetric division at the 8-cell stage and inherit a fate determining polar domain (Angerer and Angerer, 2000; Davidson et al., 1998)) and in 32-cell stage blastomeres at the animal pole (p=0.85 unpaired t-test, Figure 4A,B). Thus, cell volume, but not cell fate or developmental timing, appears to dictate spindle microtubule growth rate and consequent spindle size in cleaving P. lividus embryos, as we found for C. elegans. We conclude that this scaling principle operates across a 500-fold change in cell volume and could potentially represent an evolutionarily conserved feature of cleaving embryos.
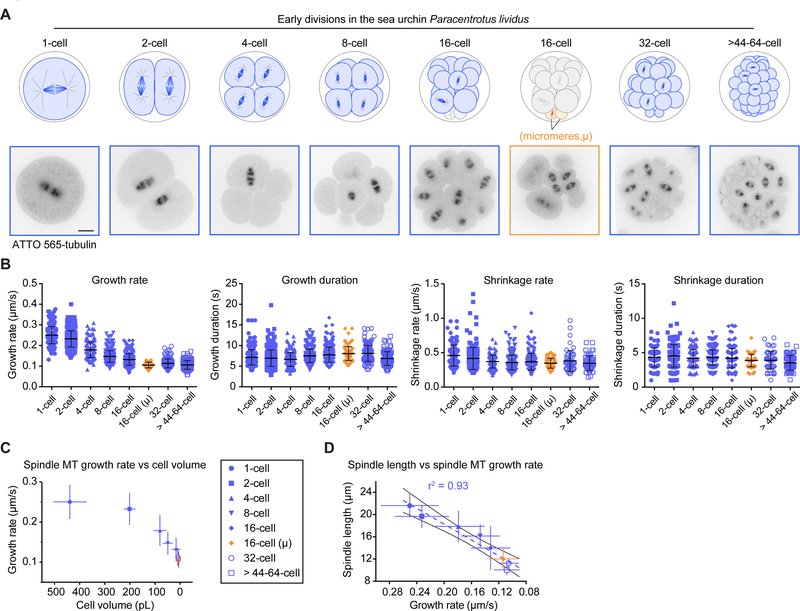
Figure 4. Spindle Length Scales with Cell Volume and Microtubule Growth Rate During Embryo Cleavage in the Sea Urchin P. lividus.
(A) (Top)(Left) Schematic representations of early embryonic divisions of the sea urchin Paracentrotus lividus. Top views schematized for the 2- and 4-cell stages. All other schemes represent side views with the animal pole at the bottom. At the 16-cell stage, micromeres highlighted in orange. (Bottom)(Right) Still frames from confocal live imaging of P. lividus embryos microinjected with ATTO 565-labelled pig brain tubulin during the first six embryonic divisions (1- to >44–64-cell stage). Scale bar, 20 μm.
(B) Microtubule dynamics parameters for spindle microtubules plotted at each cleavage stage (1- to >44–64-cell stage). Each dot corresponds to an individual microtubule. Microtubule growth rate significantly varies between stages (one-way ANOVA with Tukey’s multiple comparison tests: p<0.0001), except between the 16-cell micromeres, the 32-cell and the >44–64-cell macromeres (p>0.5 in all cases). However, the 16-cell micromeres and the 32-cell macromeres, as well as the 32-cell and the >44–64-cell macromeres differ significantly using a Student t-test (p=0.0004 and 0.0039 respectively), but the 16-cell micromeres do not when compared to the >44–64-cell macromeres (p=0.85). All parameters including sample size, mean and SD are listed in Table S2. (Horizontal bars, means; error bars, SD; n(cells/stage)≥5, n(microtubules/stage)≥55).
(C) Mean growth rate for spindle microtubules plotted at each cleavage stage (1- to >44–64-cell stage) over the average corresponding cell volume. (Error bars, SD).
(D) Mean spindle length plotted at each stage (1- to >44–64-cell stage) over the corresponding average spindle microtubule growth rate. Dotted blue line and black lines represent the linear regression and 95% confidence interval respectively. Pearson correlation coefficient (r2) is indicated at the top of the graph because p ≤ 0.01. (Error bars, SD).
3D Simulations Reproduce Spindle Length Scaling with Microtubule Growth Rate
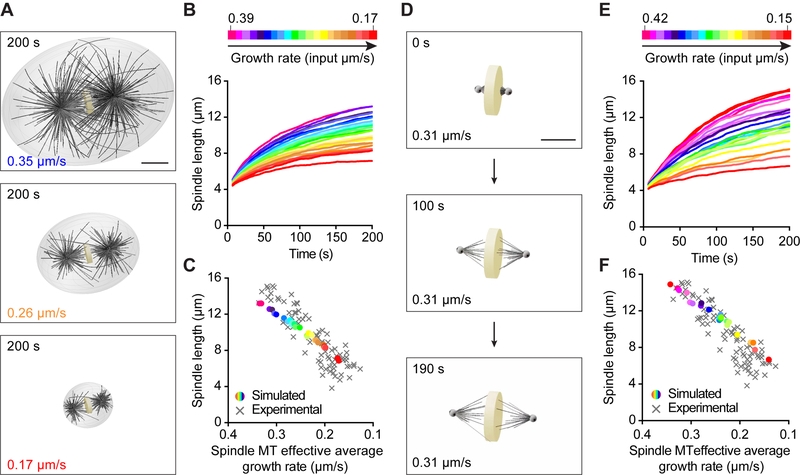
To directly test if modulating only microtubule growth rate is sufficient to explain the observed spindle length scaling, we utilized computer simulations. Using the cytoskeletal simulator Cytosim to develop a three-dimensional spindle model, we examined the contribution of the microtubule growth rate on spindle size in a dose-dependent manner in silico (www.cytosim.org; Nedelec F, 2007). We modeled each spindle pole as an aster composed of a preset number of microtubules that can grow, shrink, and stochastically undergo catastrophe and rescue (Figure 5A and S4, see detailed parameters in Table S3 and Methods). We constructed two asters by distributing a subset of these microtubules evenly and then confining the remaining microtubules within a cone to account for the higher microtubule density directed toward the metaphase plate in C. elegans spindles. We represented the metaphase plate itself using a fixed discoid volume in the center of the cell that microtubules cannot enter. Using our experimental measurements, we scaled cell volume, metaphase plate thickness and spindle width linearly with respect to the input growth rate (Table S2B). Microtubules are confined inside the cell volume, and growing plus-ends contacting the cell boundaries can create pushing forces leading to aster centration. Similarly, growing microtubules emanating from the two centrosomes, and contacting the metaphase plate, can generate forces that would push spindle poles apart leading to spindle elongation. To ensure that spindle pole movements would strictly occur through growing microtubules pushing on the cell cortex or on the metaphase plate, we omitted microtubule motors from this model. We analyzed the effects of modulating the microtubule growth rate as a function of cell volume, while keeping the three other microtubule dynamics parameters constant. We tested the physiological range of microtubule growth rates that we measured in vivo in C. elegans (Table S1). In this first set of simulations, the spindle length (pole-to-pole distance) trended toward a plateau over time for every tested microtubule growth rate (Figure 5B and Movie S3). Consistent with our hypothesis, the steady state spindle length decreased concomitantly with the microtubule growth rate (Figure 5C). Overall, the spindle lengths calculated in silico were in good agreement with our experimental measurements (Figure 5C), which confirms that this simple model accurately recapitulated the in vivo relationship between microtubule growth rate and spindle length.
Figure 5. Microtubule Growth Rate Scales Spindle Length in Computational 3D Spindle Models.
(A) Spindle length scaling with microtubule growth rate and cell volume. Astral and spindle microtubules and a cell boundary are included in these simulations. Images correspond to steady state spindles obtained after running the simulations for 200 s. The input growth rate indicated at the bottom left of each image. Scale bar, 5 μm.
(B) Simulated spindle length plotted over time at various microtubule growth rates. Color-coding of the growth rate indicated at the top, from magenta (0.39 μm/s) to red (0.17 μm/s).
(C) Simulated steady state (200 s) spindle length plotted over the effective average spindle microtubule growth rate. Growth rate color-coded as in (B). Experimental data in grey.
(D) Spindle length scaling with microtubule growth rate. Only spindle microtubules are included in these simulations. Images correspond to a spindle obtained with a microtubule growth rate of 0.31 μm/s at the beginning of the simulation (0 s, top) and after running the simulation for 100 s (middle) and 190 s (bottom). Scale bar, 5 μm.
(E) Simulated spindle length plotted over time at various effective average spindle microtubule growth rates. Color-coding of growth rate indicated at the top, from magenta (0.42 μm/s) to red (0.15 μm/s).
(F) Simulated steady state (200 s) spindle length plotted over spindle microtubule growth rate. Growth rate color-coded as in (E). Experimental data in grey.
Since our experimental data suggested that spindle length correlates with spindle but not astral microtubule dynamics, we tested if a simulation devoid of cell boundary and astral microtubules could reproduce the relationship between spindle length and microtubule growth rate (Figure 5D). With this modified model, we tested a range of microtubule growth rates, while keeping all other parameters constant (Figure 5E and Movie S4). We did find that spindle length plateaued over time at a length that increased with microtubule growth rate that yielded similar values and scaling slopes as in our experiments (Figure 5F). This model exhibited the correct multiplicative factor between <L> and spindle length, which suggests that this scaling factor only depends on the geometrical dimensions of the system (i.e. cell size and/or dimensions of the metaphase plate) included in the simulation. Together, these findings suggest that modulating spindle microtubule growth rate is sufficient to scale spindle size with cell volume in cleaving embryos.
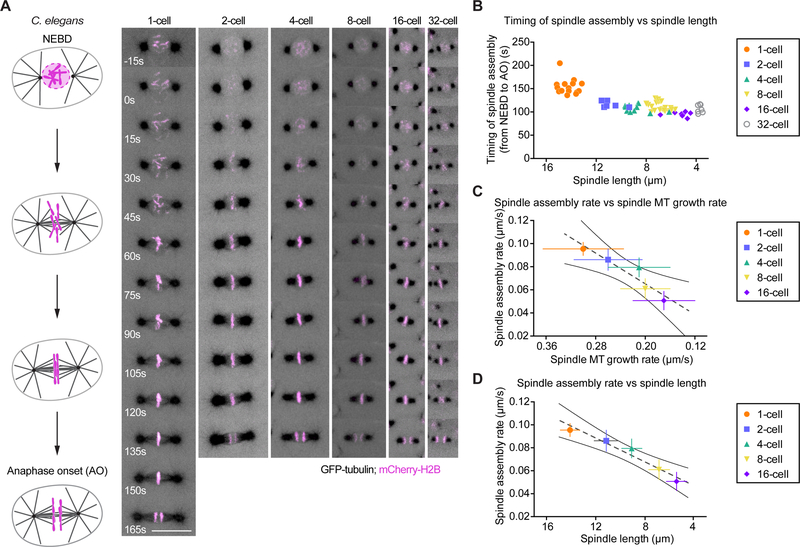
Spindle Assembly and Microtubule Growth Rates Proportionally Correlate with Constant Spindle Assembly Timing During Embryo Cleavage
An interesting finding from our 3D simulations was that spindle length reached steady state within a timeframe that was independent of microtubule growth rate or final spindle size (Figure 5B, E and S4D). So, our simulations predict that shorter spindles would not assemble faster than longer ones. To test if this occurred in vivo, we compared spindle lengths at various stages of C. elegans cleavage with the timing of spindle assembly (Figure 6A and Movie S5). Since spindle length continuously increases from NEBD to anaphase onset in C. elegans embryos, we approximated spindle assembly timing as the duration between NEBD and anaphase onset. We found that the timing of spindle assembly was independent of the initial distance between centrosomes or final spindle length (Figure 6B). Although spindle assembly took slightly longer in the 1-celled zygote than in subsequent stages, spindle assembly durations stayed relatively constant over the first five rounds of embryonic division, despite the progressive decrease in spindle length at each division. We also observed a progressive decrease of the spindle assembly rate, which strongly correlated with the decreasing spindle length and microtubule growth rate across early embryonic divisions (including in the zygote, Figure 6C and D). These results agree with the predictions from our simulations. Therefore, the progressive decrease of the microtubule growth rate is sufficient to regulate the constant timing of spindle assembly in cleaving embryos.
Figure 6. Spindle Assembly Rate Scales with Spindle Microtubule Growth Rate During C. elegans Embryo Cleavage.
(A) Left: Schematics of the spindle assembly process in the C. elegans embryo from NEBD (top) to anaphase onset (bottom). Right: Still frames from live confocal imaging of C. elegans embryos co-expressing mCherry-tagged Histone H2B (Magenta) and GFP-tagged β-tubulin (Grey) during spindle assembly in the first six embryonic divisions (1- to 32-cell stage from left to right). Timings are relative to NEBD. Images correspond to maximum projections of z-stacks. Scale bar, 10 μm.
(B) Spindle assembly timing measured from the 1- to the 32-cell stage in C. elegans embryos and plotted over the corresponding spindle length at anaphase onset. Each dot corresponds to an individual spindle. (n≥9 per stage).
(C) Mean spindle assembly rate (Spindle length/Spindle assembly timing) measured at each cleavage stage from the 1- to the 16-cell stage plotted over the corresponding average spindle microtubule growth rate. Dotted and solid grey lines represent the linear regression and 95% confidence interval respectively.
(D) Mean spindle assembly rate (Spindle length/Spindle assembly timing) measured at each cleavage stage from the 1- to the 16-cell stage plotted over the corresponding average spindle length. Dotted and solid grey lines represent the linear regression and 95% confidence interval respectively.
Discussion
In summary, we have explored the dynamic properties of microtubules as cell size decreases in early cleaving embryos. Using the nematode C. elegans and the sea urchin P. lividus, our results establish that the growth rate of spindle microtubules in both species is the primary parameter of dynamic instability that decreases proportionally to cell volume and concomitantly with spindle length. Using an in silico 3D spindle model, we demonstrate that decreasing the microtubule growth rate proportionally to cell volume is sufficient to control spindle size and recapitulates the principal features of spindle scaling observed in vivo. Our results suggest a simple mechanism to adjust spindle length with varying cell volume, as typically found in early embryonic development. Importantly, our results elucidate how spindle assembly timing is constant among different blastomere sizes in early embryos. This mechanism may be crucial to prevent chromosome segregation defects (Holmfeldt et al., 2006; Lancaster et al., 2013; Mitchison et al., 2015).
Microtubule Dynamics and Cell Volume
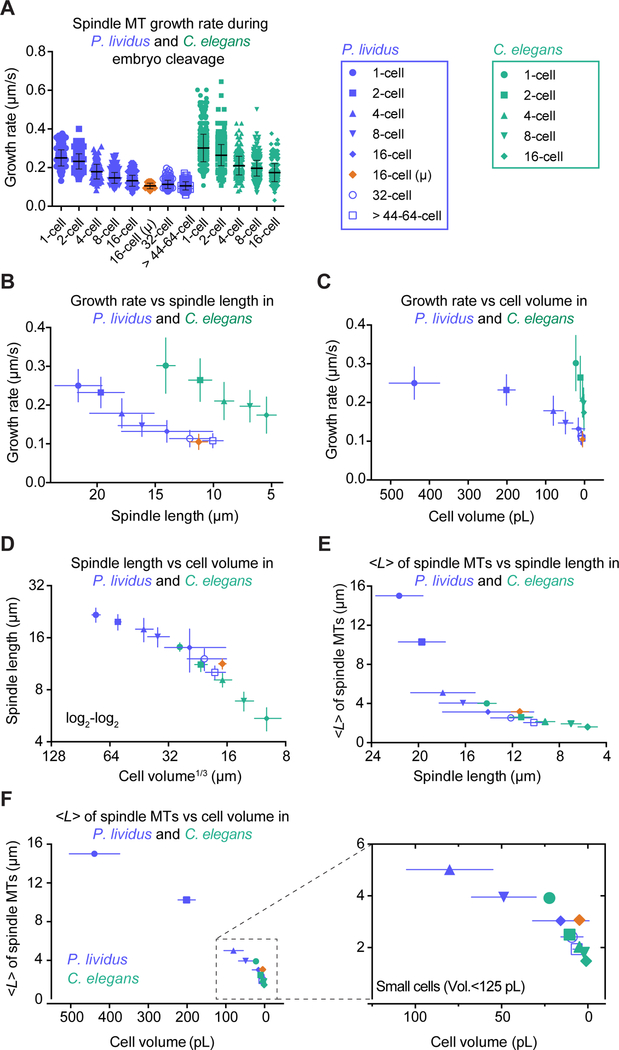
Parameters of microtubule dynamics scaled similarly across cleavages in C. elegans and P. lividus but had different absolute values in the two species. C. elegans embryos are ~20 times smaller in volume than sea urchin embryos, but their microtubules grow significantly faster at every stage of cleavage (Figure 7A). These results indicate that microtubule growth rate is not an absolute predictor of spindle length (Figure 7B–D). In contrast, the average theoretical microtubule length <L>, calculated from the four parameters of dynamic instability, followed a linear and similar trend in both species when plotted against cell volume (Figure 7F). Strikingly, blastomeres of comparable volumes displayed very similar values of <L> regardless of their species of origin (C. elegans 2-cell stage blastomeres and P. lividus 32-cell stage blastomeres display volumes of 10.6 and 9.28 pl respectively and <L> of 2.48 and 2.42 μm respectively). <L> is a readout of the overall microtubule dynamics, which suggests that microtubule dynamics vary to set an average microtubule length that scales with cell volume. Furthermore, since spindle length is roughly proportional to <L>, microtubule dynamics are modulated to scale spindle length with cell volume.
Figure 7. Microtubules and Spindle Length Adaptation to Cell Volume in P. lividus and C. elegans Embryos.
(A) Growth rate for spindle microtubules in P. lividus and C. elegans (same as Figure 1C and 4B) plotted at each cleavage stage (1- to >44–64-cell stage in P. lividus and 1- to 16-cell stage in C. elegans). Each dot corresponds to an individual microtubule. Color-coding and symbols shown in the righthand boxes. (Horizontal bars, means; error bars, SD; n(cells/stage)≥5, n(microtubules/stage)≥55) in P. lividus; n(cells/stage)≥12, n(microtubules/stage)≥288) in C. elegans. All parameters including sample size, mean and SD listed in Table S2.
(B) Mean growth rate for spindle microtubules plotted at each cleavage stage over the corresponding average spindle length. (Bars, SD).
(C) Mean growth rate for spindle microtubules plotted at each cleavage stage over the average corresponding cell volume. (Bars, SD).
(D) Mean spindle length plotted over the cube root of the average cell volume on a log2-log2 scale. (Error bars, SD).
(E) Average theoretical microtubule length <L> plotted over the experimentally measured spindle length. (Error bars, SD).
(F) (Left) Average theoretical microtubule length <L> plotted over the average cell volume. (Right) Magnification of the graph for the smaller cells (volume<125 pL). (Error bars, SD).
However, we note that <L> is not equal to the spindle half-length nor do we suggest that spindle length is solely determined by microtubule length. Spindle assembly requires other factors (Dumont and Mitchison, 2009b; Mitchison et al., 2015; Reber and Hyman, 2015; Walczak and Heald, 2008; Waters and Salmon, 1997). We also provide evidence for this premise, as <L> did not always scale linearly with spindle length in P. lividus (Figure 7E). Specifically, 1- and 2-cell stage sea urchin blastomeres deviated from the linear scaling trend followed by other cells, and assembled spindles that were shorter than predicted by values of <L>. Several non-exclusive causes could account for the discrepancy between <L> and spindle length: microtubules may bend, detach from the centrosomes or be severed (Brangwynne et al., 2006; Crowder et al., 2015; Dumont and Mitchison, 2009a; Gadde and Heald, 2004; Goshima et al., 2005a; Maiato et al., 2004; McBeath and Fujiwara, 1990; Reber and Hyman, 2015; Redemann et al., 2017; Wuhr et al., 2008). We also note that theoretically, the microtubule growth rate should regulate microtubule mass within the spindle rather than the spindle length itself (Mitchison et al., 2015; Reber et al., 2013). So, when microtubule length and spindle length scale linearly together, total microtubule mass in the spindle may also scale. But in larger blastomeres, an excess in microtubule mass, due to the increased growth rate, could be compensated either by an increase in density, or by a change in spindle width rather than length. Consistent with the later possibility and previous observations done in other species (Crowder et al., 2015; Hara and Kimura, 2013), we observed a significant change in the spindle length to width ratio and in the centrosome radius in P. lividus among cleavages, which correlated with proportionally wider spindle poles at earlier stages (Figure S3H and Table S2).
Microtubule Dynamics and Spindle Length
Our analysis revealed that the microtubule growth rate and spindle length decrease concomitantly and sub-proportionally to cell volume across reductive divisions in early C. elegans and P. lividus embryos. This contrasts with previous work comparing stage 3 and 8 Xenopus embryo extracts. These studies found no major variation in the microtubule growth rate, but a threefold increase in the microtubule catastrophe frequency correlated with smaller spindles (Wilbur and Heald, 2013). It is possible that different mechanisms may exist across phylogeny that would produce similar effects on spindle length, thus reflecting evolutionary diversity. However, we note that Xenopus blastomeres (at both stages 3 and 8) are significantly larger than C. elegans or P. lividus embryos, which could favor one spindle scaling mechanism over the other. Moreover, stage 8 blastomeres utilize a linear scaling regime, where spindle length scales linearly with cell size, while stage 3 blastomeres remain in the ‘large-cell’ regime, in which spindle length reaches an upper limit that is independent of cell size (Wilbur and Heald, 2013; Wuhr et al., 2008). The difference in the catastrophe frequency between stage 3 and 8 could thus be related to the transitioning from ‘large-cell’ to linear scaling regime rather than reflecting a cell size-dependent spindle scaling mechanism. Finally, results obtained with Xenopus egg extracts could also reflect the removal of membrane confinements. In the ‘large-cell’ regime, spindle length is controlled through spindle intrinsic mechanisms such as autocatalytic microtubule nucleation (Decker et al., 2018; Ishihara et al., 2016; Mitchison et al., 2015). It is therefore possible that in the ‘small-cell’ regime, modulation of the microtubule nucleation rate similarly contributes to spindle length scaling. However, our results demonstrate that regulation of the microtubule growth rate is sufficient to account for spindle length adaptation to cell volume in small cells.
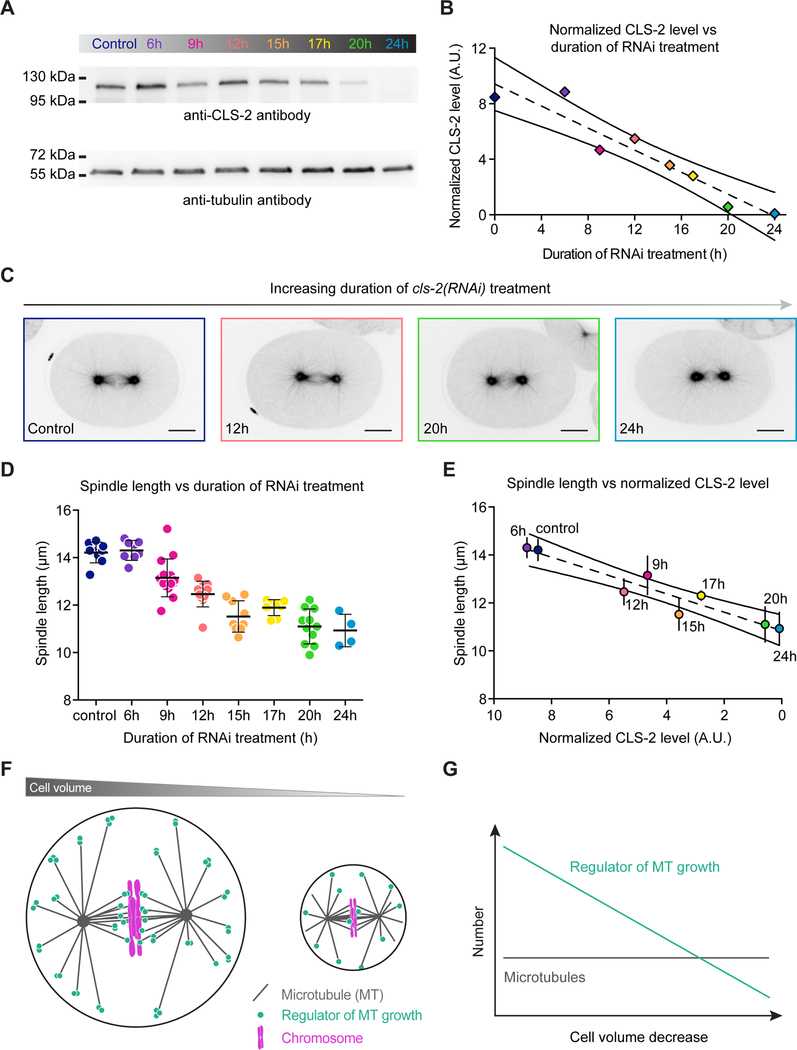
We still must determine the specific factors that control microtubule growth rate with respect to cell volume. The limiting component hypothesis proposes that one or several factors critical for spindle assembly have a limited supply, so their absolute quantity per cell diminishes upon embryo cleavage (Mitchison et al., 2015; Reber and Goehring, 2015). Tubulin is the most obvious candidate, as its concentration does determine the rate of microtubule elongation in vitro (Walker et al., 1988). However, several in vitro and in vivo studies argue against tubulin being the sole limiting component. In Xenopus egg extracts or in the presence of crowding agents in vitro, the growth rate of microtubules is not linearly dependent on the available tubulin concentration (Parsons and Salmon, 1997; Wieczorek M, 2013), as robust spindle length scaling occurred in lipid droplets even after artificially increasing the amount of tubulin (Good et al., 2013). In sea urchin embryos, a 5-fold excess of tubulin is present in a soluble non-polymerized form, and can be artificially mobilized into the spindle upon treatment with crowding agents indicating that tubulin itself is not limiting in this system (Inoue and Sato, 1967; Raff et al., 1975; Raff and Kaumeyer, 1973; Rebhun et al., 1975). Quantitative western blotting allowed us to estimate the tubulin concentration to approximately 22 μM in C. elegans embryos, which is similar to Xenopus embryos (Figure S5 and Methods) (Belmont et al., 1990). Based on a recent study providing the first complete electron tomographic reconstruction of the mitotic spindle in the C. elegans zygote (Redemann et al., 2017), we estimated that the total amount of tubulin heterodimers should be sufficient to assemble 2–3 zygotic mitotic spindles (see Methods). Thus, tubulin seems unlikely to be the only limiting factor for spindle length scaling. Another possibility is that specific microtubule-associated proteins (MAPs) that regulate microtubule growth rate are also limiting. Consistent with this, we found that reducing levels of the CLS-2 protein in C. elegans zygotes modulated microtubule growth rate and spindle length proportionally. Furthermore, by performing a temporal dilution experiment through progressive RNAi depletion of CLS-2 in the C. elegans one-cell embryo, we found that spindle length was proportional to CLS-2 protein level (Figure 8A–E). Together, these results are in agreement with the limiting amount of one or several MAPs restricting microtubule dynamics and spindle length in early embryos (Figure 8F,G). To explain how the absolute amount of MAPs could become limiting and thus control spindle length scaling, we favor a model where a progressive mismatch is established across successive divisions between the number of available MAPs and the number of spindle microtubules (Figure 8G). The CLS-2 and TPXL-1 proteins are good candidate factors for acting as limiting components (Greenan et al., 2010). However, MAPs influence several microtubule dynamics parameters, which complicates the identification of the key factor(s) that coordinates spindle length with cell size in early embryos in vivo (Bowne-Anderson et al., 2015). We must also point out that the limiting component model cannot account for the spindle-scaling mismatch observed between oocytes and zygotes of most species, and that cell volume is not an absolute predictor of spindle length. Indeed, although they assemble in an identical volume of the same cytoplasm, oocyte spindles are usually smaller than their zygotic counterpart. Meiotic-specific and volume-independent mechanisms can usually account for the discrepancy between cell volume and spindle length in oocytes (Clandinin and Mains, 1993; Clark-Maguire and Mains, 1994; Lu et al., 2004; Srayko et al., 2000).
Figure 8. A Limiting Component(s) Model for Microtubule Growth Rate and Spindle Length Scaling.
(A) (Top) Western blot using an anti-CLS-2 antibody (C2a14) on whole control worm extract or extracts treated with cls-2(RNAi) for increasing times (from 6 to 24h). Duration of the RNAi treatment is indicated above each lane. (Bottom) Western blot using an anti-α-tubulin antibody (DM1α) was used as a loading control.
(B) Normalized CLS-2 level plotted over the corresponding duration of RNAi treatment. Dotted and solid black lines represent the linear regression and 95% confidence interval respectively. The color code corresponds to (A).
(C) Still images of one-cell C. elegans embryos in metaphase expressing GFP-tagged β-tubulin after progressive depletion of the CLS-2 protein. The duration of the RNAi treatment is color-coded as in (A) and is indicated at the bottom right corner of each image. Scale bar, 10 μm.
(D) Spindle length in one-cell C. elegans embryos in metaphase after progressive depletion of the CLS-2 protein plotted over the corresponding duration of RNAi-treatment. The color code corresponds to (A). (Error bars, SD).
(E) Mean spindle length plotted over the corresponding level of CLS-2 protein. Dotted and solid black lines represent the linear regression and 95% confidence interval respectively. (Error bars, SD).
(F) A limiting component model explains microtubule growth rate and spindle length scaling with respect to cell volume (Decker et al., 2011; Goehring and Hyman, 2012; Good et al., 2013; Hazel et al., 2013; Mitchison et al., 2015; Schmoller and Skotheim, 2015). In this model, one or several positive regulators of spindle microtubule growth (such as CLS-2) bind to microtubule (+)-ends. The microtubule growth rate depends on the number of (+)-end-bound regulators. Across successive divisions, the concentration of regulators remains constant but the absolute number of regulator molecules decreases with cell volume. If in parallel, the number of spindle microtubules remains constant or decreases in a sub-proportional manner across divisions, the number of regulators becomes progressively limiting relative to the number of microtubule (+)-ends. The limited number of regulators in turn restricts spindle microtubule growth rate.
(G) Schematic evolution of spindle microtubule and regulator number as cell size decreases. As cells get smaller, microtubules are shorter but their number remains constant (or decreases slower than the number of regulators). In contrast, the absolute number of regulator molecules is directly proportional to cell volume. If the regulator stimulates microtubule growth (such as CLS-2), microtubule growth rate will progressively decrease as cell volume decreases across successive divisions.
Microtubule Dynamics and Spindle Assembly Rate
In addition to spindle size, we also found that the rate of spindle assembly (spindle length/time of assembly) scaled with cell size in C. elegans embryos. Our simulations combined with our in vivo measurements suggest that this scalability occurs through the proportional relationship between microtubule growth rate and steady state spindle length. This property could be key in early embryos that undergo rapid cell divisions with a weak spindle assembly checkpoint (Encalada et al., 2005). Indeed, C. elegans embryos respond to unattached kinetochores by inducing a small delay in mitosis but do not arrest their cell cycle progression. In this context, adapting the microtubule growth rate to the final spindle length would ensure efficient chromosome bi-orientation and avoid sister chromatid mis-segregation. Fast-assembling microtubules in large blastomeres reach further and search more space to ensure timely chromosome capture during the short mitotic time window. As cleavage progresses and spindle length decreases, slowing down microtubule assembly may negate the simultaneous connection of individual kinetochores to both spindle poles, which would promote merotely and gross chromosome mis-segregation if not corrected. From these observations, we envision that decreasing the microtubule growth rate permits the embryo to not only scale down its spindles, but also globally optimize chromosome segregation during embryogenesis. Previous work in C. elegans embryos demonstrated that the rate of spindle elongation during anaphase (Hara and Kimura, 2009) and the rate of cortical ring constriction during cytokinesis (Carvalho et al., 2009) also decreased as cell volume decreased, which makes the time needed for anaphase spindle elongation and for cytokinetic ring constriction independent of cell size. Future studies should examine whether the rates of other cell division processes also exhibit intrinsic scalability and whether this is a general principle that dictates how cell biological processes scale with cell size.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Julien Dumont (julien.dumont@ijm.fr). Requests related to the simulations should also be addressed to Francois Nedelec (nedelec@embl.de).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans
C. elegans are small, transparent hermaphroditic soil worms with rapid, invariant and fully characterized development (Sulston and Horvitz, 1977; Sulston et al., 1983). Spindle assembly has been extensively studied in C. elegans (Muller-Reichert et al., 2010), primarily because many key proteins and mechanisms are highly conserved with other metazoans. All C. elegans strains were maintained at 23°C or at 16°c for the cyk-1(ts) mutant strain (JCC49 and JDU156) and cultured using standard procedures (Brenner, 1974). Briefly, C. elegans worms were grown monoxenically on NGM plates seeded with the OP50 E. coli strain. Worm strains were propagated and maintained by transferring three L4 larvae on fresh seeded NGM plates every 3 to 6 days. Different strains were obtained either by stable single insertion by MosSCI (Frokjaer-Jensen et al., 2008) or by crossing with pre-existing strains. C. elegans worm strains and genotypes are listed in the Key Resources Table. Adult hermaphrodites were dissected for embryo collection and analysis.
Paracentrotus lividus
Paracentrotus lividus sea urchins were purchased from the Roscoff Marine Station (Station Biologique de Roscoff, CNRS-UPMC, France) or from the EMBRC-France (European Marine Biological Resource Center, Banyuls-sur-mer, France). Adult individuals were maintained in large Plexiglas tanks filled with artificial seawater at 16°C under constant oxygenation and recurrent filtering, siphoning and volume exchange. Sea urchins were exposed to photoperiods of 12h light and 12h darkness. Water constants: temperature (16°C), density (1.025 g/L), pH (8 to 8.3) and carbonate hardness (7–10 dKH) were controlled on a daily basis. Others constant such as phosphates and nitrogen were controlled on a weekly basis and maintained within range compatible with sea urchin physiology (PO4 < 0.05 mg/L; NH4 < 0.1 mg/L; NO3 ≤ 1 mg/L; NO2 < 0.025 mg/L). Sea urchins were fed with a mix of frozen algae in agar. Adult males and females were induced to spawn. P. lividus have no sexual dimorphism and sex could only be determined during spawning based on the color of the laid gametes (eggs are pink to red, sperm are white). Sex of imaged embryos was therefore not determined.
METHOD DETAILS
RNA-mediated interference (RNAi) and drug treatment in C. elegans
Depletion of C27D9.1 and of CLS-2 (R107.6) in Figure 8 were done by feeding as described by Kamath et al., (Kamath et al., 2001). For L1 larvae synchronization, adult worms were bleach-treated (1.2% NaOCl, 250 mM KOH in water; (Stiernagle, 2006) to release embryos. Embryos were allowed to hatch and left to starve in M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl). Synchronized L1 hermaphrodite larvae were grown on regular NGM plates seeded with the OP50 bacterial strain for 44 hours (until they reached the adult stage), and then adult worms were transferred on RNAi feeding plates for at least 24 hours for C27D9.1 and for an increasing time (from 6 to 24h) for CLS-2 respectively. RNAi feeding was performed on NGM plates containing 50 μg/ml ampicillin (Euromedex) and 1 mM IPTG (Isopropyl β-D-thiogalactopyranoside, Euromedex) and seeded with the HT115(DE3) bacterial strain transformed with the empty L4440 vector (control) or containing the C27D9.1 target sequence (corresponding to nt179–1366 of the genomic sequence). Depletion of CLS-2 in Figure 3 was performed by microinjection of the corresponding dsRNA in L4 hermaphrodite larvae as previously described (Fire et al., 1998). The dsRNA targeting cls-2 was synthesized from a purified PCR product (corresponding to nt164–1081 of the genomic sequence, QIAquick PCR purification, Qiagen) that contained the T3 and T7 phage RNA polymerase promoter sequences (T3: taatacgactcactatagg; T7 : aattaaccctcactaaagg) in 5’ and 3’, respectively, using the MEGAscript T3 and T7 transcription kits (ThermoFischer Scientific). Single stranded RNAs were purified using the MEGAclear Transcription Clean-Up kit (ThermoFischer Scientific). Sense and antisense single stranded RNAs were mixed in equimolar concentrations and annealed at 68°C for 10 min followed by 37°C for 30 min. DsRNAs were aliquoted, snap-frozen in liquid nitrogen and stored at −80°C until use. The dsRNA targeting cls-2 was injected in L4 hermaphrodites at a concentration of 1.8 μg/μl. Partial depletion of CLS-2 as obtained 25 to 27 h post-injection at 23°C (Maton et al., 2015). PTR-2 (C32E8.8) depletion was performed in a humid chamber by soaking 25 to 30 L4 hermaphrodite larvae in a 5 μl drop of the corresponding dsRNAs (synthesized as previously and corresponding to nt1886–2904 of the genomic sequence) supplemented with 2.86 mM spermidine (Sigma) and 0.05 % gelatin (Sigma). L4 larvae were soaked at 20°C for 24 hours, then washed twice in M9 medium and transferred on NGM plates for another 24 hours at 20°C before observation. PTR-2-depleted embryos are defective in eggshell formation and are therefore permeable and amenable to drug treatments. To induce a metaphase arrest in zygotes, PTR-2-depleted adult worms were dissected in 20 μM clasto-lactacystine β-lactone (CLβL, Merck Millipore) in L-15 blastomere culture medium [0.5 mg/mL Inulin (Sigma); 25 mM HEPES pH 7.5; 3 mL Leibowitz L-15 Media and 20% (vol/vol) Heat-Inactivated FBS (Life Technologies)] with 1% DMSO (Euromedex) final concentration.
Estimation of the tubulin heterodimer concentration in the C. elegans embryo by quantitative western blot
C. elegans early embryos (1- to ~64-cell stage) were extracted from synchronized adult N2 worms by bleaching as described earlier in the “RNA-mediated interference (RNAi) and drug treatment in C. elegans” method section. Embryos were then resuspended in M9 buffer. The number of embryos per volume and their approximate number of blastomeres (stage) were estimated under a dissection scope (Nikon). Embryos were then lysed into 1× Laemmli sample buffer (80 mM Tris-HCl pH 6.8, 140 mM 2-Mercaptoethanol, 2.5% SDS, 10% glycerol) by vortexing in presence of 0.7 mm diameter Zirconia beads (BioSpec Products) 3 times 5 min at 4°C with pauses of 5 min. Lysis efficiency was assessed using a dissection scope. Samples were then incubated 5 min at 95°C before loading on a SDS-PAGE gel. The equivalent of ~1120 or ~2240 embryos were loaded on a SDS-PAGE gel (10%, 37.5:1 acrylamide:bisacrylamide) together with increasing amounts of porcine brain tubulin purified as described in “Tubulin preparation and fluorescent labeling” (Figure S5). C. elegans and porcine brain tubulin samples were treated similarly. Proteins were transferred onto a nitrocellulose blotting membrane Protran 0.45 μm (Amersham, GE Healthcare) by liquid transfer. Membrane was incubated with DM1α antibody (Sigma) diluted 1/1000 in 5% fat-free milk in Tris-buffered saline (TBS: 50 mM Tris pH 7.5, 150 mM NaCl). Protein bands were revealed with an HRP-labeled goat anti-mouse IgG 1:10,000 (GE Healthcare) and chemoluminescence (ECL SuperSignal West Pico, ThermoScientific). Detection was performed on a luminescence image analyzer LAS-4000 (Fujifilm). Band intensities were analyzed using ImageJ. The amount of α-tubulin contained per embryo was estimated to be between 2.04 and 2.73×10−11 g. This amount corresponds to a concentration of 0.93 to 1.24 g/L (Embryo volume = 2.2×10−11 L, Figure S1 and Table S1) or 18.6 and 24.8 μM (molecular weight of α-tubulin = 50,000 g/mol). C. elegans microtubules are composed of 11 protofilaments on average (Chalfie and Thomson, 1982), and tubulin heterodimers are 8 nm long (Nogales et al., 1998). If we assume an average tubulin heterodimer concentration of 22 μM, a C. elegans embryo contains enough tubulin heterodimers to build 21.2 cm of microtubules in total. The metaphase spindle in the 1-celled zygote is composed of around ~20,000 microtubules, which are on average 4 μm long (Redemann et al., 2017). This average microtubule length also corresponds to the theoretical average microtubule length <L> calculated from the dynamics parameters in the 1-celled zygote (Table S1). Therefore, in the 1-celled zygote in metaphase, spindle microtubules reach a total length of about 8 cm (20,000 × 4 μm). We thus concluded that the C. elegans zygote contains enough tubulin heterodimers to build between 2 and 3 mitotic spindles.
Estimation of CLS-2 protein level following RNAi-mediated depletion in the C. elegans embryo by quantitative western blot
Control or RNAi treated gravid C. elegans were collected and washed into M9 buffer containing 0.1% Triton X-100. Worms (100 worms per 100 μl) were then lysed into 1× Laemmli sample buffer (80 mM Tris-HCl pH 6.8, 140 mM 2-Mercaptoethanol, 2.5% SDS, 10% glycerol) by vortexing in presence of 0.7 mm diameter Zirconia beads (BioSpec Products) 3×5 min at 4°C with pauses of 5 min. Lysis efficiency was assessed using a dissection scope. Samples were then incubated 5 min at 95°C before loading on a SDS-PAGE gel. The equivalent of 10 worms were loaded on a SDS-PAGE gel (8%, 37.5:1 acrylamide:bisacrylamide). Proteins were transferred onto a nitrocellulose blotting membrane Protran 0.45 μm (Amersham, GE Healthcare) by liquid transfer. The membrane was incubated with a rabbit polyclonal custom-made anti-CLS-2 antibody (C2a14) diluted at 1μg/ml and then with a DM1α antibody (Sigma) diluted 1/1000 in 5% fat-free milk in Tris-buffered saline (TBS: 50 mM Tris pH 7.5, 150 mM NaCl). Protein bands were revealed with an HRP-labeled goat anti-rabbit (CLS-2) or anti-mouse (DM1α) IgG 1:10,000 (GE Healthcare) and chemoluminescence (ECL Prime, Amersham). Detection was performed on a luminescence image analyzer LAS-4000 (Fujifilm). Band intensities were analyzed using ImageJ.
Tubulin preparation and fluorescent labeling
Tubulin was obtained from pig brains following cycles of polymerization and depolymerization (Castoldi and Popov, 2003). Tubulin was then labeled with NHS-ester-ATTO 565 (ATTO-TEC) and further purified through two polymerization/depolymerization cycles (Hyman, 1991). Briefly, unlabeled polymerized tubulin was incubated 40 min at 37°C in the presence of 5 mM of succinimidyl ester-coupled fluorophore in labeling buffer (0.1 M HEPES pH 8.6, 1 mM MgCl2, 1 mM EGTA and 40% glycerol (volume/volume). Microtubules were then spun down through a low pH cushion (60% glycerol BRB: 80 mM PIPES pH 6.8, 1 mM MgCl2, 1 mM EGTA) and resuspended and depolymerized in 50 mM K-glutamate pH 7.0, 0.5 mM MgCl2. Depolymerized labeled tubulin was recovered through 120,000 g centrifugation at 2°C and suspended in 4 mM MgCl2, 1 mM GTP and 1× BRB from a 5× BRB stock. Tubulin was then allowed to repolymerized at 37°C for 40 min. Microtubules were pelleted at 120,000×g for 20 min at 37°C and depolymerized in ice-cold BRB buffer. The depolymerized tubulin was recovered from a 10 min centrifugation at 2°C and at 150,000×g diluted to 15–20 mg/ml in 1× BRB, aliquoted, frozen in liquid nitrogen and stored at −80°C.
Two-photon live microscopy of C. elegans embryos
Embryo and blastomere volume measurements were made from Z-stacks of 1- to 16-cell stage embryos expressing a GFP-tagged membrane marker (OD58 strain that expresses the GFP-PH-domain under a pie-1 germline promoter). Embryos were released from gravid adult hermaphrodites in a 5 μl drop of egg salt buffer (ESB, 25 mM HEPES pH 7.3, 118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2) by cutting each worm open using a scalpel (blade N°15). Embryos were then transferred between two 3% agarose pads to prevent compression and to minimize refractive index mismatch. The montage was then placed on a glass slide, sealed with paraffin. A well of Valap (petroleum jelly:lanolin:paraffin, 1:1:1) was made around it and filled with water for imaging in immersive conditions. An upright two-photon microscope with a Zeiss Plan APO 20 7l/NA1.0 water objective was used (Lavision Biotec). GFP was excited with a pulsed laser at 920 nm. Emission at 535 nm was collected with a GaAsP detector (Lavision Biotec). The entire embryo was imaged with 1 μm Z-sectioning and 190 nm lateral sampling.
Spinning-disc live microscopy of C. elegans embryos
Gravid worms were dissected as described in the previous paragraph in egg salt buffer (ESB, see above). Embryos were transferred on a 3% agarose pad between a glass slide and a coverslip. The montage was then sealed with Valap. Live imaging was done using a spinning-disk (CSU-X1, Yokogawa) confocal microscope (Roper Scientific) coupled to a CoolSnap HQ2 CCD camera (Photometrics) and all acquisition parameters were controlled by MetaMorph 7 software (Molecular Devices). For microtubule dynamics parameter extraction, images were acquired on a single channel (491 nm excitation) every 0.5 sec with a single z-plan using a Nikon CFI APO LBDA S 60×/NA1.4 oil objective with no binning. To measure spindle assembly timings, images were acquired on two channels sequentially (491 and 561 nm excitations) every 5 sec with 11 z-plan at 1 μm intervals using a Nikon CFI APO LBDA S 40×/NA1.25 water objective with no binning. Maximal projections are presented.
Injection and Imaging of sea urchin embryos
Gametes were collected by intracoelemic injection of 0.5M KCl. Eggs were washed several times in fresh seawater, kept at 12–14°C and used within 24h. Sperm were kept concentrated for several days at 4°C. Upon use, the sperm suspension was diluted 1000 times in fresh seawater and activated by vigorous aeration. Freshly collected P. lividus eggs were dejellied by pouring through a 80 μm pore Nitex mesh (Sefar). Dejellied eggs were immobilized on a protamine (Sigma) treated 60 mm diameter glass-bottom dish for both microinjection and imaging. Eggs were fertilized 20 min after ATTO 565-tubulin microinjection (2 mg/ml) by adding a few drops of activated sperm. Live imaging was done using a spinning disk (CSU-W1, Yokogawa) confocal microscope (Leica Microsystems) coupled to an ORCA-Flash4.0 V2 CMOS camera (Hamamatsu). Embryos were maintained and imaged between 18°C and 20°C during and in between acquisitions. All acquisition parameters were controlled by MetaMorph 7 software (Molecular Devices). Images were acquired on a single channel (561 nm excitation) every 1 sec with a single z-plan using a Leica Plan APO 63×/NA1.2 water objective.
Immunostainning of sea urchin embryos
Freshly collected eggs were fertilized in a 140 mm diameter glass container and allowed to develop at 18°C (same as for live imaging). Embryo cleavage and NEBD were monitored by transmitted light on a dissection scope. At NEBD, embryos were fixed following previously described procedures (Foe and von Dassow, 2008; Tanimoto et al., 2016). Fixation was performed in bulk in 15 ml polypropylene conical tubes. Embryos were first fixed for 70 min in 100 mM Hepes, pH 6.9, 50 mM EGTA, 10 mM MgSO4, 2% formaldehyde, 0.2% glutaraldehyde, 0.2% Triton X-100, and 400 mM dextrose. They were then rinsed three times for 10 min in PBS and placed in freshly made 0.1% NaBH4 in PBS for 45 min. Embryos were rinsed again with PBS and PBS O.1% Triton X-100 (PBT). For microtubule immunostaining, embryos were incubated overnight at 4°C with a primary anti–α-tubulin antibody (DM1α, Sigma-Aldrich) at a 1:5000 dilution, and washed 3 times for 30 min in PBT. Embryos were then incubated overnight in secondary antibody (goat anti-mouse DyLight 550, Jackson ImmunoResearch) at a 1:1000 dilution in PBT. Chromatin was stained using DAPI at a 1:5000 dilution in PBS. Embryos were then mounted in 0.5% n-propyl gallate in a 9:1 solution of glycerol/1M TRIS ph8.0.
Image analysis
Two-photon acquisitions of GFP-PH (membrane marker) during C. elegans embryonic development were analyzed using Imaris software (Bitplane). C. elegans blastomere volumes were extracted after semi-automated surface reconstruction. P. lividus blastomere volumes were estimated from experimentally measured blastomere diameters. Spinning-disk confocal images were analyzed using the Fiji package (http://fiji.sc/; https://imagej.nih.gov/ij/). Spindle length was estimated as the aster-to-aster distance (Crowder et al., 2015) on GFP-tubulin images. Metaphase plate thickness and centrosome radius were measured on segmented mCherry-histone and GFP-β-tubulin images in C. elegans embryos or ATTO 565-tubulin images in P. lividus embryos. At each cleavage stage in C. elegans embryos, a 10% or 40% grey level threshold were applied on mCherry-histone or GFP-β-tubulin images respectively. In P. lividus embryos, a 30% grey level threshold was applied on ATTO 565-tubulin images for centrosome radius measurements. These thresholds were selected to obtain the sharpest limit between the metaphase plate histone signal or the centrosome and the cytoplasmic background with minimal erosion of the signal. Measured values were used in Cytosim simulations to build in silico spindle 3D models.
Analysis of microtubule dynamics
Individual embryos were imaged at a single stage of cleavage and for less than 10 min to avoid photoxicity-induced bias (Lacroix and Maddox, 2014; Lacroix et al., 2016). Kymographs were traced from time-lapse sequences using the ‘Multi kymograph’ plugin (http://imagej.net/Multi_Kymograph) in Fiji. Polymerization and depolymerization rates, as well as catastrophe and rescue frequencies, were extracted from these kymographs by manual tracking and semi-automated parameter extraction using homemade macros in Fiji and Excel (Microsoft) software. Catastrophe and rescue frequencies were defined as the inverse of the average growth time or shrinkage time, respectively (Desai and Mitchison, 1997; Komarova et al., 2002; Walker et al., 1988). Although this method could potentially overestimate catastrophe and rescue occurrences (which would be identical in all tested experimental conditions), it allows estimating both parameters independent of microtubule pausing, moving out-of-focus during live imaging or entering in the fluorescence-saturated centrosome area.
Cytosim simulations
All simulations were performed in 3D using Cytosim (www.cytosim.org; Nedelec F, 2007). Cytosim simulations have already been used to show how polymerization forces and thus microtubules dynamics could affect centrosome positioning (Letort et al., 2016). Following this approach, we aimed for a minimal model in which spindle length, corresponding to the distance between two asters, would be influenced by microtubules dynamic behavior. A detailed list of Cytosim parameters for each object in the simulations is presented in Table S3. Spindle poles were modeled using microtubules radiating radially from a sphere (asters). The surface of the sphere contains a preset number of nucleation sites, at which microtubules are anchored at their minus-ends constraining their position and orientation. The minus-ends are static, while the plus-ends may grow or shrink, stochastically switching between these states following the usual phenomenological model of dynamic instability. The metaphase plate was represented as a fixed and rigid disc positioned orthogonally at the center of the “cell”. Although microtubules were excluded from inside this plate, microtubule-associated molecules (called “singles”, each made of one “hand”) anchored on this disc and capable of tracking growing microtubule ends were used to mimic the chromokinesins present on chromosomes, as well as the capture of microtubules by pro-metaphase kinetochores. The primary function of these microtubule-associated molecules is to prevent microtubules from sliding on the surface of the “metaphase plate”. At the start of every simulation, the two asters were positioned symmetrically 2 μm away from the “metaphase plate” on each side.
The input growth rate () varied within the range of experimental measurements made in C. elegans embryos from 1- to 16-cell stage. The effective microtubule growth rate was set to decrease exponentially under an opposing force as , where is the input growth rate and is the force at which microtubule polymerization stalls (Dogterom and Yurke, 1997). Consequently, microtubules encountering the metaphase plate grew dramatically slower, in agreement with in vivo measurements at the kinetochore (Maddox et al., 2003; Tirnauer et al., 2002). Further, in the first simulation that contained both astral and spindle microtubules, a force-dependent increase in the catastrophe rate (see also Table S3) induced microtubule shrinking soon after cortical or metaphase plate contact. This force-dependent microtubule shrinking was not implemented in the second simulation (with only spindle microtubules). So, the effective average growth rate of microtubules in our simulations represented the mean rate of free growing microtubules and stalled microtubules at the metaphase plate (or at the cortex) (Figure S4A).
Two different spindle models were tested, as described below. (Figure 5A–C): This model included 500 microtubules per aster. 400 microtubules were allowed to grow uniformly in space, and 100 microtubules per aster were constrained in a cone directed toward the metaphase plate to mimic the higher microtubule density observed in vivo in the spindles of C. elegans embryos. C. elegans blastomeres were represented by 3D ellipsoids. Cell volume, metaphase plate thickness and radius were scaled together with respect to the input microtubule growth rate using realistic values extrapolated from our experimental measurements (TableS2B). Scaling to the input growth rate was done assuming a linear proportionality between microtubule growth rate and other dimensions. Due to their initial proximity to the metaphase plate, spindle microtubules were more likely to be stalled and therefore displayed a lower effective average growth rate than astral microtubules (Figure S4B). This was particularly true for higher initial rates and was reminiscent of the difference in the growth rates we measured in vivo between astral and spindle microtubules. This mild difference in the astral and spindle effective growth rates was sufficient to induce a significant deviation of the average microtubule length reached by the two populations (Figure S4C). The average length difference was not evident at low initial growth rates but was particularly clear for rates above 0.3 μm/sec. This result is in agreement with the evolution of <L> for astral and spindle microtubules calculated from in vivo measured parameters, which became closer between the two types of microtubules as cells get smaller (Table S1). The faster decrease of the average length of astral compared to spindle microtubules is likely due to increasing cortical surface to cell volume ratio as cleavage progresses. Indeed, the cortical surface decreases slower than the cell volume, so that astral microtubules are more likely to encounter the cortex and shrink, as blastomeres get smaller. Our simulation therefore accurately recapitulates two key features of the dynamic behavior of astral and spindle microtubules in vivo during embryonic cleavage: 1) spindle microtubules grow slower and are, on average, shorter than astral microtubules during early cleavages and 2) these differences tend to fade as blastomeres become smaller (Figure S4B,C).
(Figure 5D–F): 50 microtubules were constrained in a spindle-shaped cone to reflect the C. elegans embryo spindle geometry (Table S2). Importantly, increasing or decreasing the number of microtubules had no effect on the final outcome of the simulations (data not shown). The two asters do not include astral microtubules and grow in an infinite space without a cell boundary.
QUANTIFICATION AND STATISTICAL ANALYSIS
All graphs were made in Prism 6 (GraphPad Software). Statistical analyses, including Student t-tests, one-way ANOVA multiple comparisons, linear regressions and Pearson and Spearman correlation tests were performed in Prism 6 (GraphPad software). Microtubule dynamics parameters have different variances, so we used the unequal variance unpaired t-test with the Welch’s correction for normally distributed populations with unequal standard deviation. Tukey or Dunnett tests were performed for multiple comparisons following one-way ANOVA. In Figure 4B, multiplicity adjusted p value was estimated for the multiple comparison Tukey’s test.
Supplementary Material
Movie S1. Microtubule dynamics in a dividing Paracentrotus lividus embryo. Related to Figure 4. Movie from live confocal acquisitions of a 2-cell stage P. lividus embryo microinjected with ATTO 565-tubulin (grey). A single focal plane was imaged every second. Scale bar, 20 μm
Movie S2. Microtubule dynamics in a dividing Paracentrotus lividus embryo. Related to Figure 4. Movie from live confocal acquisitions of a 16-cell stage P. lividus embryo microinjected with ATTO 565-tubulin (grey). A single focal plane was imaged every second. Scale bar, 20 μm.
Movie S3. In silico assembly of a steady-state length mitotic spindle in a confined space with astral microtubules. Related to Figure 5. Movie from a simulation made with Cytosim as presented in Figure 5A–C. Input microtubule growth rate in this simulation is 0.35 μm/s. Scale bar, 10 μm
Movie S4. In silico assembly of a steady-state length mitotic spindle in an infinite space and without astral microtubules. Related to Figure 5. Movie from a simulation made with Cytosim, as presented in Figure 5D–F. Input microtubule growth rate in this simulation is 0.31 μm/s. Scale bar, 10 μm
Movie S5. Timing of spindle assembly during C. elegans embryo cleavage. Related to Figure 6. Combined stacks from live confocal imaging of C. elegans embryos co-expressing mCherry-tagged Histone H2B (Magenta) and GFP-tagged β-tubulin (Grey) during spindle assembly in the first six embryonic divisions (1- to 32-cell stage from left to right). Movies correspond to maximum intensity projection of z-stacks. Movies start 15 s prior to NEBD and end after anaphase onset. Note that spindle assembly takes slightly more time in the one-cell embryo. Scale bar, 10 μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| DM1α anti-tubulin | Sigma-Aldrich | T9026 |
| Rabbit anti-CLS-2 directed against N-term 14 amino acids of C. elegans CLS-2 (R107.6): MSRVISRSTPGGTC | Home made (purified from rabbit serum) | C2a14 |
| Bacterial and Virus Strains | ||
| OP50 E. coli | Caenorhabditis Genetics Center (CGC) | http://www.cgc.cbs.umn.edu/strain.php?id=11078 |
| HT115(DE3)E. coli | Caenorhabditis Genetics Center (CGC) | http://www.cgc.cbs.umn.edu/strain.php?id=8854 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| ATTO 565 NHS-ester | ATTO-TEC GmbH | AD 565–35 |
| Protamine sulfate | Sigma-Aldrich | P4020 |
| Clasto-Lactacystin β-Lactone (CLβL, proteasome inhibitor) | Merck Millipore | 426102 |
| 3XmCherry-EMTB | Gift from George von Dassow, University of Oregon | N/A |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Nematode Caenorhabditis elegans, strain JDU19: [5’mex-5 ::GFP::tbb-2; mCherry::his-11; cb-unc-119(+)] I; unc-119(ed3) III | Julien Dumont lab, CNRS Institut Jacques Monod, Paris | N/A |
| Nematode Caenorhabditis elegans, strain JA1559: [pJA138; 5'pie-1::mCherry::tbb-2::3'pie-1] | Julie Ahringer lab | N/A |
| Nematode Caenorhabditis elegans, strain MDX20: unc-119(ed3). ojIs1[unc-119(+) pie-1::GFP::tbb-2] | Maddox lab, University of North Carolina in Chapel Hill | N/A |
| Nematode Caenorhabditis elegans, strain JDU156 :ijmSi7 [pJD348/pSW077; 5'mex-5::GFP::tbb-2; mCherry::his-11; cb-unc-119(+)] I; cyk-1(or596ts) III; unc-119(ed3) III | Julien Dumont lab | N/A |
| Nematode Caenorhabditis elegans, strain JCC49: cyk-1(or596ts) III; ltIs37[pAA64; pie-1::mCHERRY::his-58; unc-119 (+)] IV | Julie C. Canman lab | N/A |
| Nematode Caenorhabditis elegans, strain OD58: unc-119(ed3) III; ltIs38[pAA1; pie-1::GFP::PH(PLC1delta1); unc-119 (+)] | Oegema & Desai lab | N/A |
| Nematode Caenorhabditis elegans, wild-type isolate (Bristol) | Caenorhabditis Genetics Center (CGC) | N2 |
| Sea urchin Paracentrotus lividus | Station Biologiques de Roscoff et Banyuls-sur-mer, France http://www.embrc-france.fr/fr | N/A |
| Oligonucleotides | ||
| Forward primer for production of dsRNA
targeting cls-2 R107.6:
TAATACGACTCACTATAGGttcaaggaaaagttggacc (Uppercase: T3 sequence) |
(Maton et al., 2015) | N/A |
| Reverse primer for production of dsRNA
targeting cls-2 R107.6:
AATTAACCCTCACTAAAGGggtgcatttctgattccacc (Uppercase: T7 sequence) |
(Maton et al., 2015) | N/A |
| Forward primer for production of dsRNA
targeting ptr-2 C32E8.8:
TAATACGACTCACTATAGGtgtgcccgatcattctgcat (Uppercase: T3 sequence) |
This study | N/A |
| Reverse primer for production of dsRNA
targeting ptr-2 C32E8.8:
AATTAACCCTCACTAAAGGgtggccatccaagagctgat (Uppercase: T7 sequence) |
This study | N/A |
| Forward primer for production of dsRNA against C27D9.1 using L4440 feeding vector in HT115 bacteria: tcagcaaccagcacattctc | https://www.sourcebioscience.com/products/life-science-research/clones/rnai-resources/c-elegans-rnai-collection-ahringer/ | SourceBioscience Location II-4A13 |
| Reverse primer for production of dsRNA against C27D9.1 using L4440 feeding vector in HT115 bacteria: aaaatacgcttgacgttggc | https://www.sourcebioscience.com/products/life-science-research/clones/rnai-resources/c-elegans-rnai-collection-ahringer/ | SourceBioscience Location II-4A13 |
| Forward primer for production of dsRNA against cls-2 (R107.6) using L4440 feeding vector in HT115 bacteria: aaatcgtcctcgaacattgg | https://www.sourcebioscience.com/products/life-science-research/clones/rnai-resources/c-elegans-rnai-collection-ahringer/ | SourceBioscience Location III-4J10 |
| Reverse primer for production of dsRNA against cls-2 (R107.6) using L4440 feeding vector in HT115 bacteria: tgatgggttttgacaaacga | https://www.sourcebioscience.com/products/life-science-research/clones/rnai-resources/c-elegans-rnai-collection-ahringer/ | SourceBioscience Location III-4J10 |
| Recombinant DNA | ||
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/index.html |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| GraphPad PRISM 6.0 | GraphPad | https://www.graphpad.com/ |
| Imaris | Bitplane | http://www.bitplane.com/imaris/imaris |
| Cytosim | Nédélec Laboratory | http://github.com/nedelec/cytosim |
| Other | ||
ACKNOWLEDGMENTS
We thank all members of the Dumont lab for support and advice, and Kimberley Laband for critical reading of the manuscript. We are grateful to Patricia Moussounda for providing technical support. We thank Life Science Editors for editing assistance. We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of IBiSA and France-BioImaging (ANR-10-INBS-04) infrastructures. We thank Nicolas Tissot (ImagoSeine) for his help with the 2-photon microscopy. We thank Julie Ahringer (The Gurdon Institute, University of Cambridge, UK) for the generous gift of the JA1559 C. elegans strain, and George Von Dassow, University of Oregon) for providing recombinant 3xmCherry-EMTB. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). EMBRC-France is supported by the Investments of the Future program under reference ANR-10-INBS-02. We thank Pascal Romans (UPMC, Banyuls-sur-Mer, France) and Anaëlle Pierre (IJM, Paris, France) for technical assistance with sea urchins. We thank Dmitry Ershov (IJM, Paris, France) for his help with MATLAB. We are grateful to Jean-Claude Labbe (IRIC) for initial technical support and discussions. B.L. was originally supported by a post-doctoral fellowship from the Fondation pour la Recherche Médicale (FRM ARF20140129055). JS and NM are supported by the European Research Council (CoG Forcaster N° 647073). This work was supported by CNRS and University Paris Diderot and by grants from the Agence Nationale pour la Recherche (ANR-16-CE13-0020-01), the Mairie de Paris (Emergence) and the Fondation pour la Recherche Médicale (FRM DEQ20160334869) to J.D.
Footnotes
DATA AND SOFTWARE AVAILABILITY
Data availability
All data presented in this manuscript are available upon request to the lead author (julien.dumont@ijm.fr).
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Agrell I (1956). A mitotic gradient as the cause of the early differentiation in the sea urchin embryo. Zoological Papers in Honour of B Hanstrom, pp. 27–34. [Google Scholar]
- Andersen SS (2000). Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol 10, 261–267. [DOI] [PubMed] [Google Scholar]
- Andersen SS, Ashford AJ, Tournebize R, Gavet O, Sobel A, Hyman AA, and Karsenti E (1997). Mitotic chromatin regulates phosphorylation of Stathmin/Op18. Nature 389, 640–643. [DOI] [PubMed] [Google Scholar]
- Angerer LM, and Angerer RC (2000). Animal-vegetal axis patterning mechanisms in the early sea urchin embryo. Dev Biol 218, 1–12. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, and Mitchison TJ (1990). Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell 62, 579–589. [DOI] [PubMed] [Google Scholar]
- Bowne-Anderson H, Hibbel A, and Howard J (2015). Regulation of Microtubule Growth and Catastrophe: Unifying Theory and Experiment. Trends Cell Biol 25, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, and Weitz DA (2006). Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol 173, 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, and Heald R (2007). Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol 176, 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, and Oegema K (2008). Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science 322, 1543–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A, Desai A, and Oegema K (2009). Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell 137, 926–937. [DOI] [PubMed] [Google Scholar]
- Castoldi M, and Popov AV (2003). Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif 32, 83–88. [DOI] [PubMed] [Google Scholar]
- Chalfie M, and Thomson JN (1982). Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. J Cell Biol 93, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YH, and Marshall WF (2010). Scaling properties of cell and organelle size. Organogenesis 6, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YH, and Marshall WF (2012). How cells know the size of their organelles. Science 337, 1186–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, MacLeod I, Yates JR 3rd, Oegema K, and Desai A (2005). The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol 15, 771–777. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, and Mains PE (1993). Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics 134, 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S, and Mains PE (1994). Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component. J Cell Biol 126, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder ME, Strzelecka M, Wilbur JD, Good MC, von Dassow G, and Heald R (2015). A comparative analysis of spindle morphometrics across metazoans. Curr Biol 25, 1542–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Cameron RA, and Ransick A (1998). Specification of cell fate in the sea urchin embryo: summary and some proposed mechanisms. Development 125, 3269–3290. [DOI] [PubMed] [Google Scholar]
- Davies T, Jordan SN, Chand V, Sees JA, Laband K, Carvalho AX, Shirasu-Hiza M, Kovar DR, Dumont J, and Canman JC (2014). High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev Cell 30, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker F, Oriola D, Dalton B, and Brugues J (2018). Autocatalytic microtubule nucleation determines the size and mass of Xenopus laevis egg extract spindles. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker M, Jaensch S, Pozniakovsky A, Zinke A, O’Connell KF, Zachariae W, Myers E, and Hyman AA (2011). Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol 21, 1259–1267. [DOI] [PubMed] [Google Scholar]
- Desai A, and Mitchison TJ (1997). Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13, 83–117. [DOI] [PubMed] [Google Scholar]
- Dogterom M, and Yurke B (1997). Measurement of the force-velocity relation for growing microtubules. Science 278, 856–860. [DOI] [PubMed] [Google Scholar]
- Dumont S, and Mitchison TJ (2009a). Compression regulates mitotic spindle length by a mechanochemical switch at the poles. Curr Biol 19, 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, and Mitchison TJ (2009b). Force and length in the mitotic spindle. Curr Biol 19, R749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada SE, Willis J, Lyczak R, and Bowerman B (2005). A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol Biol Cell 16, 1056–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu EB, Krueger LE, Ye A, and Rose LS (2012). CLASPs function redundantly to regulate astral microtubules in the C. elegans embryo. Dev Biol 368, 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, and Bulinski JC (1999). E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci 112 (Pt 23), 4243–4255. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, and Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Foe VE, and von Dassow G (2008). Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J Cell Biol 183, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, and Jorgensen EM (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40, 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadde S, and Heald R (2004). Mechanisms and molecules of the mitotic spindle. Curr Biol 14, R797–805. [DOI] [PubMed] [Google Scholar]
- Goehring NW, and Hyman AA (2012). Organelle growth control through limiting pools of cytoplasmic components. Curr Biol 22, R330–339. [DOI] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, and Heald R (2013). Cytoplasmic volume modulates spindle size during embryogenesis. Science 342, 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, and Vale RD (2005a). Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol 171, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, and Vale RD (2005b). Length control of the metaphase spindle. Curr Biol 15, 1979–1988. [DOI] [PubMed] [Google Scholar]
- Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Julicher F, and Hyman AA (2010). Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr Biol 20, 353–358. [DOI] [PubMed] [Google Scholar]
- Hara Y, and Kimura A (2009). Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr Biol 19, 1549–1554. [DOI] [PubMed] [Google Scholar]
- Hara Y, and Kimura A (2013). An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos. Mol Biol Cell 24, 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, and Gatlin JC (2013). Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342, 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt P, Brannstrom K, Stenmark S, and Gullberg M (2006). Aneugenic activity of Op18/stathmin is potentiated by the somatic Q18-->e mutation in leukemic cells. Mol Biol Cell 17, 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA (1991). Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J Cell Sci Suppl 14, 125–127. [DOI] [PubMed] [Google Scholar]
- Inoue S, and Sato H (1967). Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J Gen Physiol 50, Suppl:259–292. [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Korolev KS, and Mitchison TJ (2016). Physical basis of large microtubule aster growth. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, and Ahringer J (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, and Mitchison T (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell 45, 329–342. [DOI] [PubMed] [Google Scholar]
- Komarova YA, Vorobjev IA, and Borisy GG (2002). Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J Cell Sci 115, 3527–3539. [DOI] [PubMed] [Google Scholar]
- Labbe JC, McCarthy EK, and Goldstein B (2004). The forces that position a mitotic spindle asymmetrically are tethered until after the time of spindle assembly. J Cell Biol 167, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, and Maddox AS (2014). Microtubule dynamics followed through cell differentiation and tissue biogenesis in C. elegans. Worm 3, e967611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Ryan J, Dumont J, Maddox PS, and Maddox AS (2016). Identification of microtubule growth deceleration and its regulation by conserved and novel proteins. Mol Biol Cell 27, 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, and Baum B (2013). Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell 25, 270–283. [DOI] [PubMed] [Google Scholar]
- Letort G, Nedelec F, Blanchoin L, and Thery M (2016). Centrosome centering and decentering by microtubule network rearrangement. Mol Biol Cell 27, 2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, and Heald R (2012). Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol 28, 113–135. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, and Heald R (2011). Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Srayko M, and Mains PE (2004). The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol Biol Cell 15, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P, Straight A, Coughlin P, Mitchison TJ, and Salmon ED (2003). Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol 162, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, and Khodjakov A (2004). Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 167, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF (2015a). How Cells Measure Length on Subcellular Scales. Trends Cell Biol 25, 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF (2015b). Subcellular size. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maton G, Edwards F, Lacroix B, Stefanutti M, Laband K, Lieury T, Kim T, Espeut J, Canman JC, and Dumont J (2015). Kinetochore components are required for central spindle assembly. Nat Cell Biol 17, 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath E, and Fujiwara K (1990). Microtubule detachment from the microtubule-organizing center as a key event in the complete turnover of microtubules in cells. Eur J Cell Biol 52, 1–16. [PubMed] [Google Scholar]
- Mitchison T, and Kirschner M (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Ishihara K, Nguyen P, and Wuhr M (2015). Size Scaling of Microtubule Assemblies in Early Xenopus Embryos. Cold Spring Harb Perspect Biol 7, a019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T, Greenan G, O’Toole E, and Srayko M (2010). The elegans of spindle assembly. Cell Mol Life Sci 67, 2195–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J (2005). PAR proteins and the establishment of cell polarity during C. elegans development. Bioessays 27, 126–135. [DOI] [PubMed] [Google Scholar]
- Nedelec FFD (2007). Collective Langevin dynamics of flexible cytoskeletal fibers. New J Phys 9, 427–427. [Google Scholar]
- Nogales E, Wolf SG, and Downing KH (1998). Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391, 199–203. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH, Stumpff J, and Su TT (2004). Embryonic cleavage cycles: how is a mouse like a fly? Curr Biol 14, R35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi E, Filosa S, De Petrocellis B, and Monroy A (1978). The pattern of cell division in the early development of the sea urchin. Paracentrotus lividus. Dev Biol 65, 38–49. [DOI] [PubMed] [Google Scholar]
- Parsons SF, and Salmon ED (1997). Microtubule assembly in clarified Xenopus egg extracts. Cell Motil Cytoskeleton 36, 1–11. [DOI] [PubMed] [Google Scholar]
- Picone R, Ren X, Ivanovitch KD, Clarke JD, McKendry RA, and Baum B (2010). A polarised population of dynamic microtubules mediates homeostatic length control in animal cells. PLoS Biol 8, e1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff RA, Brandis JW, Green LH, Kaumeyer JF, and Raff EC (1975). Microtubule protein pools in early development. Ann N Y Acad Sci 253, 304–317. [DOI] [PubMed] [Google Scholar]
- Raff RA, and Kaumeyer JF (1973). Soluble microtubule proteins of the sea urchin embryo: partial characterization of the proteins and behavior of the pool in early development. Dev Biol 32, 309–320. [DOI] [PubMed] [Google Scholar]
- Reber S, and Goehring NW (2015). Intracellular Scaling Mechanisms. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber S, and Hyman AA (2015). Emergent Properties of the Metaphase Spindle. Cold Spring Harb Perspect Biol 7, a015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, and Julicher F (2013). XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol 15, 1116–1122. [DOI] [PubMed] [Google Scholar]
- Rebhun LI, Jemiolo D, Ivy N, Mellon M, and Nath J (1975). Regulation of the in vivo mitotic apparatus by glycols and metabolic inhibitors. Ann N Y Acad Sci 253, 362–377. [DOI] [PubMed] [Google Scholar]
- Redemann S, Baumgart J, Lindow N, Shelley M, Nazockdast E, Kratz A, Prohaska S, Brugues J, Furthauer S, and Muller-Reichert T (2017). C. elegans chromosomes connect to centrosomes by anchoring into the spindle network. Nat Commun 8, 15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose LS, and Kemphues KJ (1998). Early patterning of the C. elegans embryo. Annu Rev Genet 32, 521–545. [DOI] [PubMed] [Google Scholar]
- Schmoller KM, and Skotheim JM (2015). The Biosynthetic Basis of Cell Size Control. Trends Cell Biol 25, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469. [DOI] [PubMed] [Google Scholar]
- Srayko M, Buster DW, Bazirgan OA, McNally FJ, and Mains PE (2000). MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev 14, 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, and Hyman AA (2005). Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell 9, 223–236. [DOI] [PubMed] [Google Scholar]
- Stiernagle T (2006). Maintenance of C. elegans. WormBook, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, and Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56, 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Tanimoto H, Kimura A, and Minc N (2016). Shape-motion relationships of centering microtubule asters. J Cell Biol 212, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Canman JC, Salmon ED, and Mitchison TJ (2002). EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell 13, 4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, and Hyman AA (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, and Leibler S (1992). Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol 118, 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Labbe JC, Doree M, and Karsenti E (1990). Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature 343, 233–238. [DOI] [PubMed] [Google Scholar]
- Walczak CE, and Heald R (2008). Mechanisms of mitotic spindle assembly and function. Int Rev Cytol 265, 111–158. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, and Salmon ED (1988). Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol 107, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, and Salmon E (1997). Pathways of spindle assembly. Curr Opin Cell Biol 9, 37–43. [DOI] [PubMed] [Google Scholar]
- West GB, and Brown JH (2005). The origin of allometric scaling laws in biology from genomes to ecosystems: towards a quantitative unifying theory of biological structure and organization. J Exp Biol 208, 1575–1592. [DOI] [PubMed] [Google Scholar]
- Wieczorek MCS, Brouhard GJ (2013). Macromolecular Crowding Pushes Catalyzed Microtubule Growth to Near the Theoretical Limit. Cellular and Molecular Bioengineering 6, 383–392. [Google Scholar]
- Wilbur JD, and Heald R (2013). Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. Elife 2, e00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, and Mitchison TJ (2008). Evidence for an upper limit to mitotic spindle length. Curr Biol 18, 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Microtubule dynamics in a dividing Paracentrotus lividus embryo. Related to Figure 4. Movie from live confocal acquisitions of a 2-cell stage P. lividus embryo microinjected with ATTO 565-tubulin (grey). A single focal plane was imaged every second. Scale bar, 20 μm
Movie S2. Microtubule dynamics in a dividing Paracentrotus lividus embryo. Related to Figure 4. Movie from live confocal acquisitions of a 16-cell stage P. lividus embryo microinjected with ATTO 565-tubulin (grey). A single focal plane was imaged every second. Scale bar, 20 μm.
Movie S3. In silico assembly of a steady-state length mitotic spindle in a confined space with astral microtubules. Related to Figure 5. Movie from a simulation made with Cytosim as presented in Figure 5A–C. Input microtubule growth rate in this simulation is 0.35 μm/s. Scale bar, 10 μm
Movie S4. In silico assembly of a steady-state length mitotic spindle in an infinite space and without astral microtubules. Related to Figure 5. Movie from a simulation made with Cytosim, as presented in Figure 5D–F. Input microtubule growth rate in this simulation is 0.31 μm/s. Scale bar, 10 μm
Movie S5. Timing of spindle assembly during C. elegans embryo cleavage. Related to Figure 6. Combined stacks from live confocal imaging of C. elegans embryos co-expressing mCherry-tagged Histone H2B (Magenta) and GFP-tagged β-tubulin (Grey) during spindle assembly in the first six embryonic divisions (1- to 32-cell stage from left to right). Movies correspond to maximum intensity projection of z-stacks. Movies start 15 s prior to NEBD and end after anaphase onset. Note that spindle assembly takes slightly more time in the one-cell embryo. Scale bar, 10 μm.