Fig. 1.
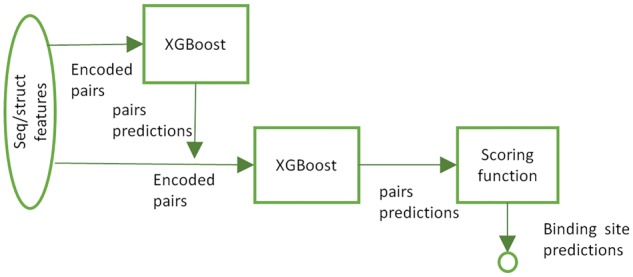
BIPSPI workflow. Sequence-base and structural features are used to codify pairs of residues. At first step, XGBoost classifier is fed with encoded pairs in order to obtain interacting pairs predictions. Interacting pairs scores are combined with original features and fed to a second step classifier. Lastly, interacting predictions obtained in step two are converted to binding site predictions employing our scoring function
