Abstract
Human norovirus (NoV) is a leading cause of fresh produce associated outbreaks. Previous research indicates that the roots of growing leafy greens and berries internalize human NoV. However the effect of plant type and inoculum level on internalization rates has not been directly compared. In this study we compared the internalization and dissemination rates of human NoV and its surrogate, Tulane virus (TV) in green onion, radishes, and Romaine lettuce. We also evaluated the effect inoculum level and plant growth matrix on the rate of viral internalization. In the hydroponic growth system, we detected internalization and dissemination of human NoV RNA in green onions. In hydroponically growing green onions inoculated with high titer TV, we found higher rates of internalization and dissemination compared to green onions inoculated with low titer TV. In soil growth systems, no infectious TV was detected in either green onion or radishes. However, in Romaine lettuce plants grown in soil approximately 4 log10 PFU/g was recovered from all tissues on day 14 p.i. Overall, we found that the type of plant, growth matrix, and the inoculum level influences the internalization and dissemination of human NoV and TV.
Keywords: Human norovirus, Tulane virus, Internalization, Fresh produce, Green onion, Radish
1. Introduction
Human norovirus (NoV) is one of the major causes of acute gastroenteritis worldwide (Allwood et al., 2004; Atmar and Estes, 2006; Dicaprio et al., 2013; Glass et al., 2009; Robilotti et al., 2015). The virus is a leading cause of foodborne illness in the United States, causing nearly 58% of foodborne disease (Dicaprio et al., 2013; Scallan et al., 2011). Fresh produce has been identified as a leading cause of foodborne illness and is a major high risk food associated with human NoV outbreaks in the US (Hall et al., 2013; Li et al., 2012; Lynch et al., 2009). Vegetable row crops (such as leafy greens) and fruits were responsible for 30% and 21%, respectively, of human NoV foodborne outbreaks in the US (2009–2012) (Hall et al., 2012).Vegetables and crops can be contaminated with human NoV at any point from farm to fork. Evidence has shown that the source of pre-harvest contamination mainly comes from soil, fertilizer, or irrigation water. Though human NoV is a major contributor to fresh produce associated outbreaks, the modes of contamination and persistence of the virus in vegetables remains poorly understood.
Human NoV is a non-enveloped single stranded positive sense RNA virus of the family Caliciviridae (Robilotti et al., 2015). The major challenge in human NoV research is that currently there is no robust cell culture system for the virus. Recently, two cell culture systems were developed for human NoV, but have not yet been optimized to produce high viral titers (Ettayebi et al., 2016; Jones et al., 2015). Therefore, much of the understanding of human NoV molecular biology, pathogenesis, and environmental stability has come from the study of surrogate viruses (Li et al., 2012; Richards, 2012). Many viruses within the family Caliciviridae have been utilized as human NoV surrogates, including murine norovirus (MNV), feline calicivirus (FCV), and Tulane virus (TV) (Cannon et al., 2006; Cromeans et al., 2014; Farkas, 2015; Hirneisen and Kniel, 2013b; Kniel, 2014; Li et al., 2012). Tulane virus (TV) is a newly recognized surrogate for human NoV and is member of the genus Recovirus within Caliciviridae (Farkas et al., 2008). TV was shown to have similar pH stability to human NoV and other surrogates at ranges from pH 3 to pH 8 (Hirneisen and Kniel, 2013b). TV causes enteric infection in primates and also recognizes histo-blood group antigens as a cellular attachment receptor, similar to human NoV (Farkas et al., 2010).
Internalization of pathogens in growing produce is considered one of the potential routes for contamination of fresh produce (Erickson, 2012; Heaton and Jones, 2008; Warriner et al., 2003; Warriner and Namvar, 2010). Bacterial pathogens, such as Salmonella enterica serovar Typhimurium and Esherichia coli O157:H7, have been shown to be internalized in vegetables including lettuce, radishes, alfalfa and green onions (Erickson et al., 2010a,b; Erickson et al., 2010a,b; Franz et al., 2007; Ge et al., 2012, 2014; Golberg et al., 2011; Hora et al., 2005; Jablasone et al., 2005; Kroupitski et al., 2009; Mitra et al., 2009; Pu et al., 2009; Sharma et al., 2009; Warriner et al., 2003). Human NoV has also been shown to be internalized in growing produce such as lettuce, spinach, and green onion (DiCaprio et al., 2015a,b; Dicaprio et al., 2012; DiCaprio et al., 2015a,b; Esseili et al., 2012; Hirneisen and Kniel, 2013a; Wei et al., 2010, 2011). However, few studies exist where multiple types of produce have been evaluated for viral internalization in parallel. In addition, the influence of initial inoculum levels on viral internalization in different types of produce has not been evaluated extensively. In this study, green onion, radishes, and Romaine lettuce were selected to evaluate the effect of growth matrix, inoculum level, and vegetable type on the internalization of human NoV and TV in fresh produce.
2. Materials and methods
2.1. Viruses and cell culture
Human NoV GII.4 strain 707 (GenBank accession number JQ798158) was originally isolated from an outbreak of acute gastroenteritis in Ohio. The virus genomic RNA was quantified by reverse transcriptase quantitative PCR (RT-qPCR) and then stored at −80 °C. TV was generously provided by Xi Jiang at Cincinnati Children’s Hospital. TV was propagated in confluent monolayers of the monkey kidney cell line MK2-LLC (ATCC, Manassas, VA). MK2-LLC cells were cultured at 37 °C in a 5% CO2 atmosphere in low-serum Eagle’s minimum essential medium (Opti-MEM; Invitrogen, Carlsbad, CA) supplemented with 2% FBS. Before virus infection, MK2-LLC cells were washed with Hanks’ balanced salt solution (HBSS) and subsequently infected with TV at an MOI of 1. After 1 h of incubation at 37 °C, 18 ml of Opti-MEM with 2% FBS was added. The virus was harvested 2 days post infection by three freeze thaw cycles followed by centrifugation at 3000 × g for 10 min to remove cell debris. After centrifugation, the supernatant was collected and virus stocks were stored at −80 °C. TV titer was determined by plaque assay.
2.2. Virus enumeration by plaque assay
TV was quantified by plaque assay in LLC-MK2 cells by seeding the cells into 6-well plates (Corning Life Sciences, Wilkes-Barre, PA) at a density of approximately 3 × 106 cells per well. After one day incubation, cell monolayers were infected with 400 μl of a 10-fold dilution series of samples, respectively, and the plates were incubated for 1 h at 37 °C with gentle agitation every 10 min. The cells were then overlaid with 2.5 ml of overlay medium (Eagle’s minimum essential medium with 1% agarose, 2% FBS, 1% sodium bicarbonate, 15 mM HEPES, and 100 units/ml penicillin, kanamycin, and streptomycin) (Invitrogen). After incubation at 37 °C for 2 days, the cells were fixed with 10% formaldehyde (Fisher Scientific, Waltman, MA) overnight. The plaques were visualized by staining with 0.05% (wt/vol) crystal violet (Sigma-Aldrich, St. Louis, MO).
2.3. Quantification of human NoV by RT-qPCR
Human NoV RT-qPCR was carried out as previously described (22). Total RNA was extracted from 100 μl of sample using an RNeasy kit (Qiagen, Hilden, Germany), followed by RT-qPCR. Firststrand cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen) using the primer VP1-P1 (5′-TTATAATACACGTCTGCGC CC-3′), which targets the VP1 gene of human NoV. The VP1 gene was then quantified by real-time PCR using custom primers and TaqMan probe (Forward primer, 5′-CACCGCCGGGAAAATCA-3′, reverse primer, 5′-GCCTTCAGTTGGGAAATTTGG-3′, and reporter, 5′-FAM- ATTTGCAGCAGTCCC NFQ-3′) on a StepOne real-time PCR machine (Applied Biosystems, Foster City, CA). TV RNA was also quantified by RT-qPCR. First strand cDNA was synthesized by SuperScriptase III (Invitrogen) using the primer TVRT (5′-AATTCCACCTTCAACCCAAGTG −3′), which targets the VP1 gene of Tulane virus. The VP1 gene was then quantified by real-time PCR using custom primers and TaqMan probe (Forward primer: 5′-TTGCAGGAGGGTTTCAAGATG-3′) (Reverse primer: 5′-CACGGTTTCATTGTCCCCATA-3′) (Probe:5′-FAM-TGATGCACACATGTGGGA-NFQ-3′). PCR and cycling parameters followed the manufacturer’s protocol (Invitrogen). Briefly, TaqMan Fast Universal Master Mix was used for all reactions. For cycling parameters, a holding stage at 95 °C was maintained for 20 s prior to cycling, followed by 50 cycles of 95 °C for 1 s for annealing and 60 °C for 20 s for extension. A standard plasmid for human NoV and TV was constructed by inserting the sequence of entire ORF2 (VP1 gene) into pGEM T-easy vector (Promega, Madison, WI). The purified plasmids for human NoV and TV with known concentration was then ten-fold serial diluted to generate a standard curve for RT-qPCR. Standard curves and StepOne software version 2.1 were used to quantify genomic RNA copies. Viral RNA was expressed as the mean log10 genomic RNA copies/ml ± standard deviation.
2.4. Plant cultivation
Seeds of green onion (Allium fistulosum L.) and radish (Raphanus sativus) were purchased from Livingston Seed (Columbus, Ohio) and stored at 4 °C. Before sowing, the seeds were soaked for 1 day at room temperature (20 °C) in a 500 ml beaker of tap water. Subsequently, the water was drained and the seeds were covered with 2 layers of wet Kimwipes (Fisher Scientific, USA) for 2 days to accelerate seed germination. After germination, the seeds were planted in 4 inch plastic pots and grown in plant growth chamber under long day conditions (22 °C with 16/8 light/dark photoperiod) in Metro-Mix® 300 soil (Sun Gro Horticulture, USA).
Two month old green onions and Romaine lettuce and one month old radishes were used for experiments. For growth in the hydroponic system, plants were removed from soil and roots were washed thoroughly with tap water to remove soil and then placed in racks above feed water. Feed water was aerated using a pump and antibiotics (100 units/ml penicillin, kanamycin, and streptomycin) were added to limit microbial growth. The feed water was not replaced during the study period. After viral inoculation, only the roots contacted the feed water and a plastic barrier separated the aerial plant tissues and feed water to limit cross contamination.
2.5. Viral inoculation and sample collection for hydroponically grown green onion and radish
For green onion grown hydroponically, TV was diluted in feed water to result in a high titer (1.0 × 106 PFU/ml) and low titer (1.0 × 104 PFU/ml) virus concentration in the feed water. Human NoV was diluted in the feed water of hydroponically growing green onions to achieve 1.0 × 105 RNA copy/ml. For hydroponically grown radishes, only the high concentration (1.0 × 106 PFU/ml) of TV was tested. For each treatment group, a 10 ml sample feed water was collected before plants were inserted and was used a control to determine the stability of TV in feed water without plants. In addition, uninoculated control groups were set up for each treatment group. The total volume of the hydroponic feed water following virus dilution was 400 ml and the initial virus titer was determined using plaque assay (TV) or RT-qPCR (human NoV). Negative control plants were kept in separate tanks and no virus was added.
At days 0 (before viral inoculation), 1, 3, 7, and 14, four plants were harvested for each condition and a sample of the feed water was collected. To minimize cross contamination, the upper most leaves were harvested first, followed by shoots, and roots were harvested last (Fig. 1A, Fig. 1B). Harvested tissues were placed in individual sterile plastic bags and each tissue was weighed. Next, the surface of each tissue was decontaminated by submersion in 1000 ppm chlorine followed by rinsing with water and inactivation of residual chlorine with 0.25 M sodium thiosulfate. Samples were then homogenized by grinding with mortars and pestles with 5 ml PBS (pH 7.0). Sample homogenates were transferred to 15 ml tubes and centrifuged at 3000 × g for 10 min to remove cellular debris and the virus-containing supernatant was then transferred to a new collection tube. No virus concentration step was performed prior to detection. Detection of viruses in plant tissue samples and feed water samples was determined by RT-qPCR (human NoV) and plaque assay (TV). The detection limit for RT-qPCR was determined to be 5 RNA copy/ml using dilution of plasmid standard in each sample matrix. The detection limit for plaque assay was determined to be 10 PFU/ml by diluting TV stock in each sample matrix. Three technical replicates were executed during RT-qPCR analysis and two technical replicates were used for plaque assay for each sample.
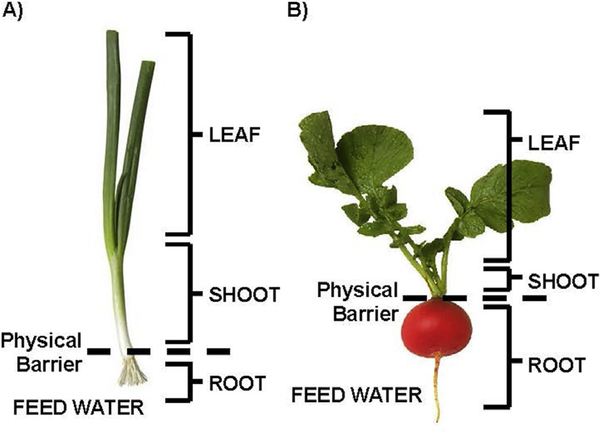
Fig. 1. Schematic representation of harvested tissues of green onion (A) and radish (B).
Leaf tissue represents the aerial tissues of the starting 2 inches above the root juncture and was harvested first. Shoot tissue represents the 2 inch portion of the aerial tissue connected to the root juncture and was not in contact with the feed water, which was harvested second. Root tissue consists of the bulb and all roots and was in direct contact with the feed water and was harvested third. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.6. Viral inoculation and sample collection for soil grown green onion, radish, and Romaine lettuce
20 ml of 1.0 × 106 PFU/ml TV was applied to the root zone of either green onion, radish, or Romaine lettuce grown in soil. The virus solution was inoculated slowly into the soil 0.5 cm from the soil surface to limit the contamination of shoot and leaves. The surface of the soil was covered with plastic film to limit viral cross contamination. The sample collection procedure was the same as described above, with the exception of the harvesting of the plant root. After harvesting leaves and shoots, roots were washed with tap water to remove soil debris. All harvested tissues were surface decontaminated with 1000 ppm chlorine and the remainder of the tissue processing was the same as described above.
2.7. Plant sap collection and virus inoculation
Growing green onions and radishes were divided into three portions, leaves, shoots, and roots, as previously described. Each portion was subjected to tissue sap (fluid) extraction by a hydraulic plant dap press (Spectrum, Plainfield, IL USA) following the manufacturer’s procedures. From each tissue, 5 ml tissue fluids were collected separately in a 15 ml tube and TV was added to the sap from each tissue to achieve at viral titer of 1 × 106 PFU/ml. At days 1, 3, 7, and 14, the virus titer in each sap was determined by plaque assay. As a positive control, TV suspended in PBS was also tested in the same manner.
2.8. Statistical analysis
Data is presented as mean ± 1 standard deviation (SD). Statistical analysis by One-way ANOVA was performed by SPSS 19 (IBM, USA). Least significant difference (LSD) was used to compare the difference of treatments. P-values less than 0.05 were considered statistically significant.
3. Results
3.1. Internalization and dissemination of human NoV in hydroponically grown green onions
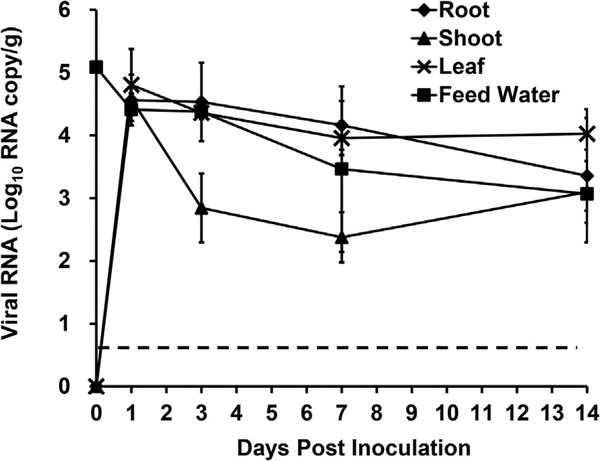
Human NoV was inoculated to the feed water of hydroponically growing green onions at a concentration of 1.0 × 105 RNA copies/ml and plants were harvested at days 1, 3, 7, and 14 post inoculation. Human NoV RNA was detected in all green onion tissues on all study days. The level of human NoV RNA in the root was the highest on day 1 post inoculation (6.0 × 104 RNA copies/g) and decreased on each following treatment day. On day 14, the virus RNA in the roots was reduced significantly compared to days 1, 3, and 7 to 4.2 × 103 RNA copies/g (P < 0.05) (Fig. 2). Similar to the results of the roots, the highest level of human NoV RNA in the shoots was detected on day 1 post inoculation and decreased on days 3, 7, and 14. The level of human NoV RNA on day 1 post inoculation was 5.9 × 104 RNA copy/g (Fig. 2). The level of human NoV RNA was reduced significantly at days 3, 7 and 14 post inoculation compared to day 1–1.8 × 103, 3.2 × 102,1.8 × 103 RNA copy/g, respectively (P < 0.05) (Fig. 2). The highest level of human NoV RNA detected in the leaves was also on day 1 post inoculation at a level of 4.6 × 104 RNA copy/g (Fig. 2). Compared to the other tissues, the level of human NoV RNA in the leaves was more stable from days 1–14 post inoculation. On days 7 and 14 post inoculation the level of human NoV RNA in the leaves was 1.0 × 104 and 1.4 × 104, respectively (Fig. 2).
Fig. 2. Human NoV internalization in hydroponically grown green onion.
Internalization kinetics plot was determined by RT-qPCR and results are reported as log10 RNA copies/g. Data points were the averages of six replicates. Error bars represent ±1 standard deviation. Dotted line represents the LOD.
The level of human NoV RNA in the feed water used to grow the green onions hydroponically had a significant reduction from 1 × 105 RNA copy/ml to 2.9 × 104 RNA copy/ml on day 1 post inoculation (P < 0.05). The level of human NoV RNA in the feed water continued to decrease over the study period and on day 14 post inoculation the RNA level was 1.3 × 103 RNA copy/ml.
3.2. Internalization and dissemination of TV in hydroponically grown green onions and radishes
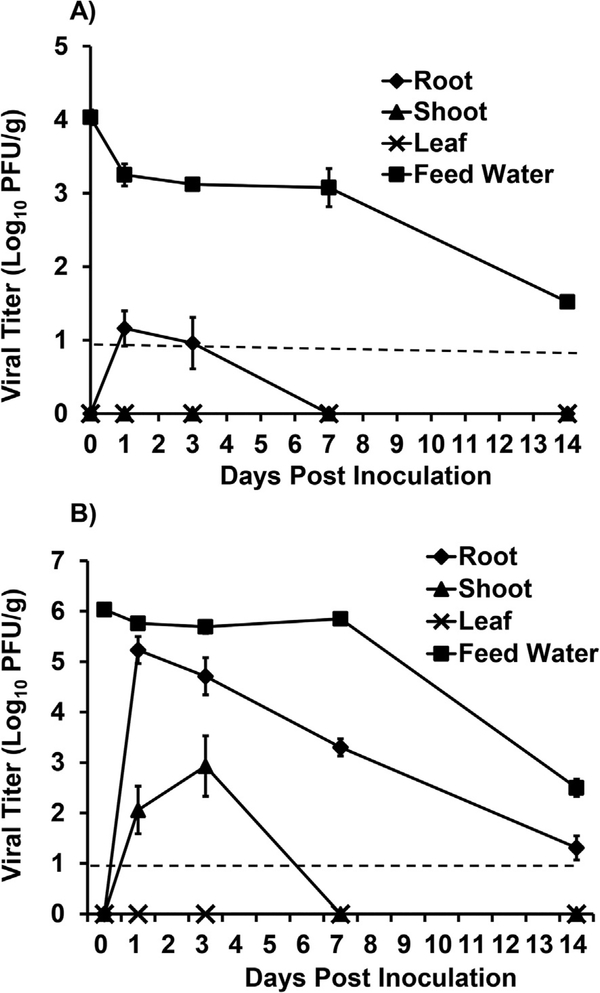
Two different starting titers of TV in the feed water for hydroponically growing green onions were tested being high, 1.0 × 106, and low, 1.0 × 104 PFU/ml. For the plants in the low titer TV treatment group, the highest TV titer detected was in the roots on day 1 post inoculation at a level of 1.6 × 101 PFU/g (Fig. 3A). The titer in the roots decreased on day 3 post inoculation to 1.1 × 101 PFU/g and on days 7, 14 post inoculation the level of virus decreased significantly and was below the detection limit (BDL) (P < 0.05). There was no detection of infectious TV in any of the leaf or shoot tissues harvested at any time-point. There was a slight decrease (<1 log) in the level of TV detected in the feed water on day 1 post inoculation and the infectious titer remained stable at this level up to day 7. There was an approximate 2.5 log10 PFU/ml reduction in the feed water TV titer on day 14 post inoculation. In the control feed water samples (without plants), the titer was slightly higher (0.5–1 log10 PFU/ml) compared to the feed water used to grow the green onions at all time-points.
Fig. 3. TV internalization in hydroponically grown green onions given low (A) and high (B) inoculum.
Viral titer is reported as logio PFU/g. Data points were the averages of six replicates. Error bars represent ±1 standard deviation. Dotted line represents the LOD.
In the hydroponically growing green onions which received the high titer TV treatment, the highest level of internalized virus detection was in the roots on day 1 post inoculation at 1.9 × 105 PFU/g (Fig. 3B). The level of TV in the roots decreased significantly at each subsequent time point (P < 0.05). On days 3, 7, and 14 the titer of TV detected in the roots was 6.9 × 104, 2.1 × 103, 2.3 × 101 PFU/g, respectively (Fig. 4). In the shoots of green onions in the high level TV treatment group, the virus was detected at day 1 at a level of 1.7 × 102 PFU/g. On day 3 post inoculation the titer in the shoots increased significantly (P < 0.05) to 1.3 × 103 PFU/g (Fig. 3B). The level of TV in the shoots was BDL on days 7 and 14 post inoculation. No infectious TV was detected in the green onion leaves at any time point. In the feed water of high titer TV treatments, there was an approximate 3.6 log10 PFU/ml reduction in the feed water on day 14 post inoculation compared to day 0. In the control feed water samples (without plants), there was no significant reduction in TV titer over the 14 day study period (P > 0.05).
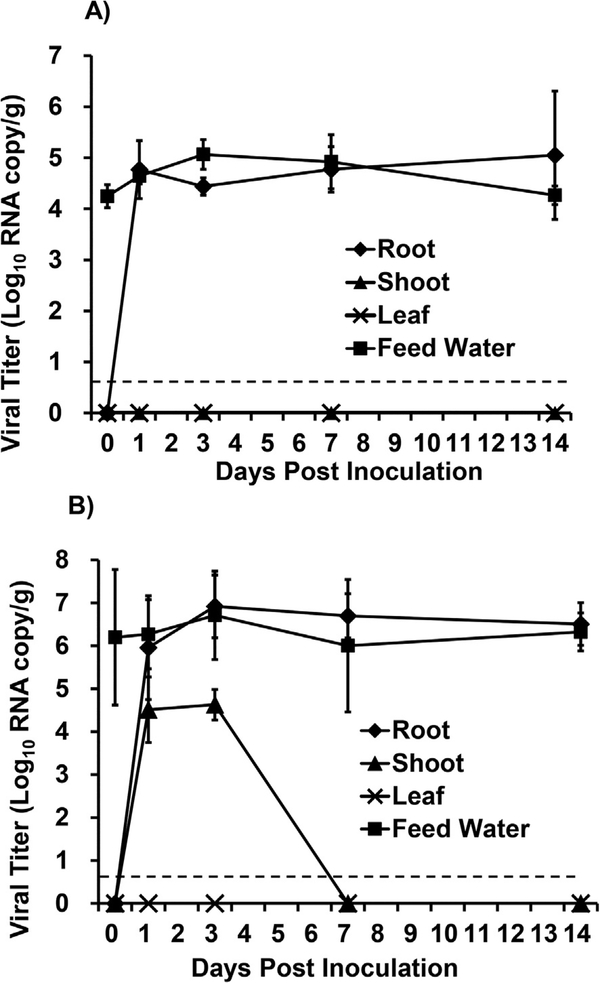
Fig. 4. TV RNA detection in hydroponically grown green onion given low (A) and high (B) inoculum.
Internalization kinetics plot was determined by RT-qPCR and results are reported as log10 RNA copies/g. Data points were the averages of six replicates. Error bars represent ±1 standard deviation. Dotted line represents the LOD.
RT-qPCR was used to determine the level of TV RNA in harvested green onion samples. The green onions inoculated with a low dose of TV had detectable RNA in the roots on day 1 post inoculation at level of 5.8 × 104 RNA copy/g (Fig. 4A). This high level of RNA remained detectable in the roots for the duration of the study (Fig. 4A). There was no TV RNA detected in either the shoots or the leaves of green onions inoculated with a low dose of TV. When green onions were inoculated with the high dose of TV there was also a high level of TV RNA detected in the roots on day 1 post inoculation 9.0 × 105 RNA copies/g and remained at this level until day 14 post inoculation (Fig. 4B). There was also TV RNA detected in the shoots of green onions inoculated with the high dose of TV on days 1 and 3 post inoculation at levels of 3.2 × 104 and 4.2 × 104 RNA copies/g, respectively. However, there was no RNA detected in the shoots at days 7 or 14 post inoculation. No TV RNA was recovered from the leaves of green onions inoculated with a high dose of TV.
Hydroponically growing radish plants also had the feed water contaminated with a high titer of TV. In contrast to green onion, there was no infectious TV recovered from any radish tissue at any time point in the study. In the feed water used to grow the radish plants, there was no reduction in infectious TV titers between day 0 and day 14 post inoculation.
3.3. TV internalization and dissemination in soil grown green onions and radishes
The soil of growing green onion plants was inoculated with 1.0 × 106 PFU of TV and tissues were harvested on days 1, 3, 7, and 14 post inoculation. TV was not detected in any green onion tissue on any day post inoculation. Soil growing radish plants were similarly inoculated with TV with the same lack of recovery of infectious TV from radish tissues.
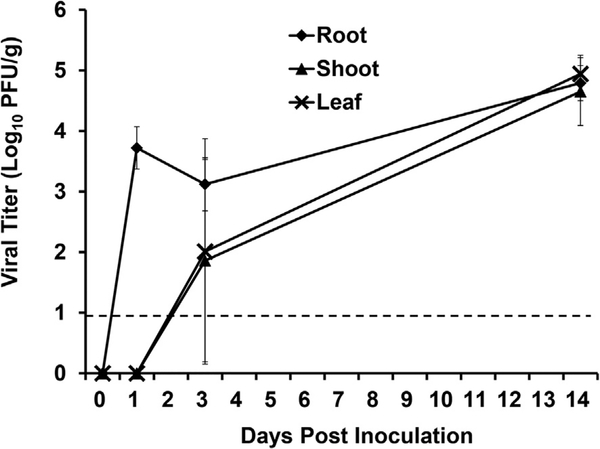
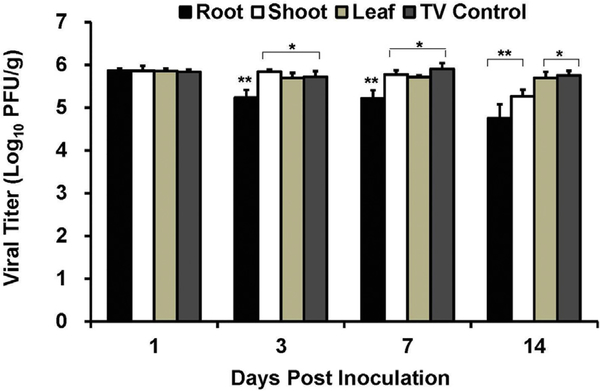
3.4. TV internalization in Romaine lettuce plants grown in soil
The soil of growing Romaine lettuce plants was inoculated with 1.0 × 106 PFU of TV and tissues were harvested on days 1, 3, and 14 post inoculation. At day 1 post inoculation, TV was detected in the roots of lettuce plants at a level of 5.24 × 103 PFU/g. No infectious TV was detected in the shoots or leaves at day 1 post inoculation (Fig. 5). On day 3 post inoculation TV was recovered in the shoots (7.2 × 101 PFU/g) and leaves (1.0 × 102 PFU/g) of Romaine lettuce, as well as the roots (Fig. 5). All harvested tissues were positive for TV on day 14 post inoculation with 6.1 × 104 PFU/g in roots, 4.4 × 104 PFU/g in shoots, and 8.7 × 104 PFU/g in leaves (Fig. 5).
Fig. 5. TV internalization in soil grown Romaine lettuce.
Viral titer is reported as log10 PFU/g. Data points were the averages of six replicates. Error bars represent ±1 standard deviation. Dotted line represents the LOD.
3.5. Stability of TV in green onion and radish saps
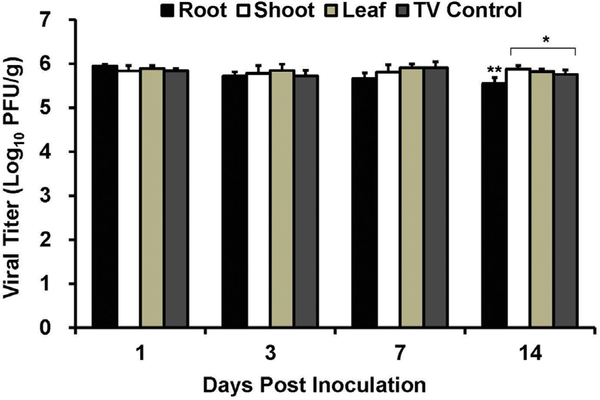
Tissue sap from the leaves, shoots, and roots of both green onions and radishes were prepared and the stability of TV in these saps was tested over 14 days. TV was stable in all green onion saps tested with no significant decreases in infectious TV titers over the 14-day study period (Fig. 6). TV was also stable in radish saps obtained from leaves and shoots (Fig. 7). However, in the root sap obtained from radishes the titer of TV was significantly decreased compared to controls on days 3, 7, and 14 (P < 0.05) (Fig. 7).
Fig. 6. Stability of TV in green onion sap.
TV was diluted 1:10 in green onion sap from root, shoot, and leaf to an approximate titer of 1 × 106 PFU/ml. TV inoculated green onion sap was stored at room temperature for 14 days, and the survival of TV was determined at day 0,1,3, 7, and 14 post inoculation by standard plaque assay. Each data point represents the average of six replicates ± standard deviation. ** denotes that values are significantly different (P < 0.01) compared to TV control. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7. Stability of TV in radish sap.
TV was diluted 1:10 in radish sap from root, shoot, and leaf to an approximate titer of 1 × 106 PFU/ml. TV inoculated green onion sap was stored at room temperature for 14 days, and the survival of TV was determined at day 0,1, 3, 7, and 14 post inoculation by standard plaque assay. Each data point represents the average of three replicates ± standard deviation. ** denotes that values are significantly different (P < 0.01) compared to TV control. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The consumption of fresh fruits and vegetables continues to increase in many countries and fresh produce is an important vehicle for foodborne disease transmission. The outbreaks associated with fresh produce have resulted in considerable public health and economic burdens (Moore et al., 2015). Among the pathogens associated with fresh produce outbreaks, an increased incidence of infections and outbreaks are attributed to foodborne viruses, most notably human NoV (Baert et al., 2011). It has been demonstrated that various types of plants are susceptible to internalization of human viral pathogens during growth (DiCaprio et al., 2015a,b; Dicaprio et al., 2012; DiCaprio et al., 2015a,b; Hirneisen and Kniel, 2013a). Various factors have the potential to affect the ability of human pathogens to internalize in growing plants including the growth substrate (soil vs. hydroponic solution), plant developmental stage, pathogen type, inoculum level, plant species and cultivar, abiotic and biotic stresses (DiCaprio et al., 2015a,b; Dicaprio et al., 2012; DiCaprio et al., 2015; Hirneisen and Kniel, 2013a). In this study, different plants types, inoculum levels, and growth matrices were tested to determine their effect on the internalization and dissemination of human NoV.
Previous research has shown that the level of pathogen inoculum used to contaminate plants can greatly influence the rate of bacterial internalization and dissemination (Pu et al., 2009). For example, in Arabidoposis thaliana, low inoculum levels (104 CFU/ml) of Salmonella Typhimurium were able to colonize the root surface but did not invade the lateral root junctions. When the inoculum level was increased to 106 CFU/mL, the bacteria were able to internalize into the root (Cooley et al., 2003). In this study, two different levels of TV inoculum, high (1 × 106 PFU/ml) and low (1 × 104 PFU/ml), were used to contaminate the feed water of hydroponically growing green onions. TV was detected in roots of green onions in both the high and low inoculum groups. The high inoculum group had infectious virus detected in the aerial portions of the plant while no virus was detected in these tissues when a low virus inoculum was applied.
Unlike bacteria that have the ability to replicate in the phyllosphere, viruses are unable to replicate outside of their host cell. However, in general it would be expected that higher inoculum levels would result in higher levels of internalization (Hirneisen and Kniel, 2013a). This was the conclusion made in another study in which higher inoculums of E. coli O157:H7 in spinach showed higher contamination incidences (Pu et al., 2009). In another study, the internalization of Salmonella Typhimurium was highly dependent on the concentration of the pathogen present in irrigation water. Irrigation water containing 5 Log CFU/mL Salmonella Typhimurium resulted in limited incidence of internalization in lettuce, while 8 Log CFU/mL Salmonella Typhimurium in the irrigation water, significantly increased the internalization frequency. Similar to these studies, we found that increased TV contamination levels in green onions lead to increased internalization of the virus in hydroponically grown green onion.
In addition, we evaluated the internalization of human NoV in hydroponically growing green onion. We found that high levels of human NoV RNA were detected in all harvested green onion tissues starting on day 1 post inoculation. The levels of RNA decreased over the study period; however RNA was still detectable in all tissues on day 14 post inoculation. The data indicates that human NoV internalized via the root of the green onions and transports to the leaves.
We also used RT-qPCR to evaluate the level of internalized TV RNA in this study. We found that high levels of TV RNA were detected starting on day 1 post inoculation and that the level of RNA remained at this level at all subsequent time points. However, this differs from the internalization results obtained for TV using plaque assay as the method of detection, in which lower levels of internalization were detected and there were reductions in the level of virus internalized at later time points. This could be due to the fact that RT-qPCR is more sensitive than plaque assay and that RNA detection alone may not accurately reflect the recovery of infectious viruses in these samples. In previous studies in hydroponically growing Romaine lettuce, it was shown that the levels of internalized TV RNA was higher than the levels of infectious virus that could be recovered from the same samples (Dicaprio et al., 2012). In addition it was found that employing an RNAse pre-treatment to degrade viral RNA that was not found in an intact viral capsid reduced the level of TV RNA detection (Dicaprio et al., 2012). Similarly, in studies investigating murine norovirus (MNV-1) and hepatitis A virus (HAV) internalization in green onions and spinach more samples were found to be positive for viral RNA compared to infectious virus (Hirneisen and Kniel, 2013a).
Two types of growth substrates, soil and hydroponic solution are commonly used in pathogen internalization studies conducted with plants. Pathogen internalization has been detected more often in hydroponically grown plants compared to plants grown in soil (Dicaprio et al., 2012). In this study, both hydroponic and soil growing green onions were contaminated with TV at a level of 1 × 106 PFU/ml. Similar to previous studies, we found that internalization of TV via the root of hydroponically growing green onion occurred while no TV internalization occurred in soil grown green onions.
The same phenomena was also observed with HAV and MNV, which were not detected in green onion plants grown in soil for up to 20 days (Hirneisen and Kniel, 2013a). Compared to soil systems, a drastic increase in virus internalization was observed in hydroponic systems with both HAV and MNV internalized up to 4 log RT-qPCR units (Hirneisen and Kniel, 2013a). Most investigators have suggested that the motility in hydroponic solution may provide more opportunity for uptake and internalization into leafy green plants compared to soil systems (DiCaprio et al., 2015a,b). The virus capsid has an ionic charge and it may remain bound to charged particles in the soil matrix reducing viral uptake by the roots (DiCaprio et al., 2015a,b). It is also possible that root damage induced by transplanting of seedlings into the hydroponic system also increased internalization in this study and others (Dicaprio et al., 2012; Franz et al., 2007).
The type of plant has also been shown to influence the rate of bacterial pathogen internalization (Erickson, 2012; Ge et al., 2012; Golberg et al., 2011; Hirneisen and Kniel, 2013a). In this study, no virus internalization occurred in radishes while there was viral internalization detected in green onions and Romaine lettuce. Internalization of Salmonella Typhimurium was also observed in lettuce and radish but not cress or spinach seedlings (Jablasone et al., 2005). In the same study, radish seedlings were susceptible to Salmonella internalization while mature radishes were resistant, indicating the importance of plant age on pathogen internalization and dissemination (Jablasone et al., 2005). The radishes used in this study were mature (30 days old). In our study we found no significant decrease in the level of virus detected in the feed water of hydroponically growing radishes. This indicates that no viral uptake occurred via radish root and it is possible that these mature radishes were not permissive to pathogen internalization.
Previously it was shown TV was internalized and disseminated in growing strawberry plants while the virus was not detected in peppers of growing green pepper plants (DiCaprio et al., 2015a,b). It was found that the green peppers had a significant anti-viral effect on TV and that this may play a role in the lack of internalization in bell pepper plants (DiCaprio et al., 2015a,b). In this study, we found that the levels of TV detected in Romaine lettuce increased over the study period, which is indicative of internalization and dissemination with no inhibition. However, the internalization and dissemination kinetics was different in green onions with high levels of internalized virus detected on day 1 post inoculation and decreasing over the study period. This may indicate that an inhibitory compound in the green onion may be inactivating the virus. Green onions contain a wide array of phenolic compounds which have the potential to act as antimicrobials (Siddiq et al., 2013). We found that sap from the green onion root lead to a 1-log reduction in infectious TV only after 14 days of exposure.
Previously, it was found that a component of radishes, trans-4-methylthio-3-butenyl isothiocyanate (TMBI), possessed antimicrobial activity (Park et al., 2016). In addition, the pigment found in the skin of radishes, anthocyanins, have also been shown to have antiviral activity. Different cyanidin glycosides present in radishes have antiviral activity against influenza A and B viruses and herpes-1 virus (Knox et al., 2003; Suzutani et al., 2003). Our data indicates that radish roots are not permissive to virus penetration and internalization; therefore, we investigated whether radishes had antiviral properties. A maximum 1-log reduction in infectious TV titer was achieved after 14 days of exposure of TV to radish sap, indicating only minimal antiviral effects.
In conclusion, we have shown that i) human NoV and TV can be internalized in hydroponically grown green onions but not in soil grown green onions, ii) the magnitude of TV internalization is influenced by the level of virus present, and iii) different types of plants have different susceptibility for viral internalization.
Acknowledgement
This work is in part supported by an internal grant from Huzhou University, P. R. China and a grant from the National Institute of Food and Agriculture, U.S. Department of Agriculture (USDA) (grant 2011-68003-30005).
Abbreviations
- NoV
norovirus
- TV
Tulane virus
- MNV
murine norovirus
- FCV
feline calicivirus
- CPE
cytopathic effect
- HBGAs
histo-blood group antigens
- DMEM
Dulbecco’s modified Eagle medium
- FBS
fetal bovine serum
- MOI
multiplicity of infection
- Opti-MEM
low serum Eagle’s minimum essential medium
- HBSS
Hank’s balanced salt solution
- RT-PCR
reverse transcriptase polymerase chain reaction
- RT-qPCR
real time reverse transcriptase polymerase chain reaction
References
- Allwood PB, Malik YS, Maherchandani S, Vought K, Johnson LA, Braymen C, Hedberg CW, Goyal SM, 2004. Occurrence of Escherichia coli, noroviruses, and F-specific coliphages in fresh market-ready produce. J. Food Prot 67, 2387–2390. [DOI] [PubMed] [Google Scholar]
- Atmar RL, Estes MK, 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am 35, 275–290 [DOI] [PubMed] [Google Scholar]
- Baert L, Mattison K, Loisy-Hamon F, Harlow J, Martyres A, Lebeau B, Stals A, Van Coillie E, Herman L, Uyttendaele M, 2011. Review: norovirus prevalence in Belgian, Canadian and French fresh produce: a threat to human health? Int. J. Food Microbiol 151, 261–269. [DOI] [PubMed] [Google Scholar]
- Cannon JL, Papafragkou E, Park GW, Osborne J, Jaykus LA, Vinje J, 2006. Surrogates for the study of norovirus stability and inactivation in the environment: aA comparison of murine norovirus and feline calicivirus. J. Food Prot 69, 2761–2765. [DOI] [PubMed] [Google Scholar]
- Cooley MB, Miller WG, Mandrell RE, 2003. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157 : H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol 69, 4915–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromeans T, Park GW, Costantini V, Lee D, Wang Q, Farkas T, Lee A, Vinje J, 2014. Comprehensive comparison of cultivable norovirus surrogates in response to different inactivation and disinfection treatments. Appl. Environ. Microbiol 80, 5743–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCaprio E, Culbertson D, Li J, 2015a. Evidence of the internalization of animal caliciviruses via the roots of growing strawberry plants and dissemination to the fruit. Appl. Environ. Microbiol 81, 2727–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicaprio E, Ma Y, Hughes J, Li J, 2013. Epidemiology, prevention, and control of the number one foodborne illness: human norovirus. Infect. Dis. Clin. North Am 27, 651–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicaprio E, Ma Y, Purgianto A, Hughes J, Li J, 2012. Internalization and dissemination of human norovirus and animal caliciviruses in hydroponically grown romaine lettuce. Appl. Environ. Microbiol 78, 6143–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCaprio E, Purgianto A, Li J, 2015b. Effects of abiotic and biotic stresses on the internalization and dissemination of human norovirus surrogates in growing romaine lettuce. Appl. Environ. Microbiol 81, 4791–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson MC, 2012. Internalization of fresh produce by foodborne pathogens. Annu. Rev. Food Sci. Technol 3, 283–310. [DOI] [PubMed] [Google Scholar]
- Erickson MC, Liao J, Payton AS, Riley DG, Webb CC, Davey LE, Kimbrel S, Ma L, Zhang G, Flitcroft I, Doyle MP, Beuchat LR, 2010a. Preharvest internalization of Escherichia coli O157:H7 into lettuce leaves, as affected by insect and physical damage. J. Food Prot 73, 1809–1816. [DOI] [PubMed] [Google Scholar]
- Erickson MC, Webb CC, Diaz-Perez JC, Phatak SC, Silvoy JJ, Davey L, Payton AS, Liao J, Ma L, Doyle MP, 2010b. Infrequent internalization of Escherichia coli O157:H7 into field-grown leafy greens. J. Food Prot 73, 500–506. [DOI] [PubMed] [Google Scholar]
- Esseili MA, Wang Q, Zhang Z, Saif LJ, 2012. Internalization of sapovirus, a surrogate for norovirus, in romaine lettuce and the effect of lettuce latex on virus infectivity. Appl. Environ. Microbiol 78, 6271–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK, 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T, 2015. Rhesus enteric calicivirus surrogate model for human norovirus gastroenteritis. J. Gen. Virol 96, 1504–1514. [DOI] [PubMed] [Google Scholar]
- Farkas T, Cross RW, Hargitt E 3rd, Lerche NW, Morrow AL, Sestak K, 2010. Genetic diversity and histo-blood group antigen interactions of rhesus enteric caliciviruses. J. Virol 84, 8617–8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T, Sestak K, Wei C, Jiang X, 2008. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol 82, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E, Visser AA, Van Diepeningen AD, Klerks MM, Termorshuizen AJ, van Bruggen AH, 2007. Quantification of contamination of lettuce by GFP-expressing Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium. Food Microbiol. 24, 106–112. [DOI] [PubMed] [Google Scholar]
- Ge C, Lee C, Nangle E, Li J, Gardner D, Kleinhenz M, Lee J, 2014. Impact of phytopathogen infection and extreme weather stress on internalization of Salmonella Typhimurium in lettuce. Int. J. Food Microbiol 168–169, 24–31. [DOI] [PubMed] [Google Scholar]
- Ge CT, Lee C, Lee J, 2012. The impact of extreme weather events on Salmonella internalization in lettuce and green onion (vol. 42, pg 1118, 2012). Food Res. Int 47,124–125. [Google Scholar]
- Glass RI, Parashar UD, Estes MK, 2009. Norovirus gastroenteritis. N. Engl. J. Med 361, 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golberg D, Kroupitski Y, Belausov E, Pinto R, Sela S, 2011. Salmonella Typhimurium internalization is variable in leafy vegetables and fresh herbs. Int. J. Food Microbiol 145, 250–257. [DOI] [PubMed] [Google Scholar]
- Hall AJ, Eisenbart VG, Etingue AL, Gould LH, Lopman BA, Parashar UD, 2012. Epidemiology of foodborne norovirus outbreaks, United States, 2001–2008. Emerg. Infect. Dis 18, 1566–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, Vinje J, Parashar UD, 2013. Norovirus disease in the United States. Emerg. Infect. Dis 19,1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton JC, Jones K, 2008. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: a review. J. Appl. Microbiol 104, 613–626. [DOI] [PubMed] [Google Scholar]
- Hirneisen KA, Kniel KE, 2013a. Comparative uptake of enteric viruses into spinach and green onions. Food Environ. Virology 5, 24–34. [DOI] [PubMed] [Google Scholar]
- Hirneisen KA, Kniel KE, 2013b. Comparing human norovirus surrogates: murine norovirus and Tulane virus. J. Food Prot 76, 139–143. [DOI] [PubMed] [Google Scholar]
- Hora R, Warriner K, Shelp BJ, Griffiths MW, 2005. Internalization of Escherichia coli O157:H7 following biological and mechanical disruption of growing spinach plants. J. Food Prot 68, 2506–2509. [DOI] [PubMed] [Google Scholar]
- Jablasone J, Warriner K, Griffiths M, 2005. Interactions of Escherichia coli O157 : 147, Salmonella typhimurium and Listeria monocytogenes plants cultivated in a gnotobiotic system. Int. J. Food Microbiol 99, 7–18. [DOI] [PubMed] [Google Scholar]
- Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, Freiden P, Graves CL, Koopmans M, Wallet SM, Tibbetts SA, Schultz-Cherry S, Wobus CE, Vinje J, Karst SM, 2015. Human norovirus culture in B cells. Nat. Protoc 10,1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniel KE, 2014. The makings of a good human norovirus surrogate. Curr. Opin. Virol 4, 85–90. [DOI] [PubMed] [Google Scholar]
- Knox YM, Suzutani T, Yosida I, Azuma M, 2003. Anti-influenza virus activity of crude extract of Ribes nigrum L. Phytother Res. 17,120–122. [DOI] [PubMed] [Google Scholar]
- Kroupitski Y, Golberg D, Belausov E, Pinto R, Swartzberg D, Granot D, Sela S, 2009. Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol 75, 6076–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Predmore A, Divers E, Lou F, 2012. New interventions against human norovirus: progress, opportunities, and challenges. Annu. Rev. Food Sci. Technol 3, 331–352. [DOI] [PubMed] [Google Scholar]
- Lynch MF, Tauxe RV, Hedberg CW, 2009. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect 137, 307–315. [DOI] [PubMed] [Google Scholar]
- Mitra R, Cuesta-Alonso E, Wayadande A, Talley J, Gilliland S, Fletcher J, 2009. Effect of route of introduction and host cultivar on the colonization, internalization, and movement of the human pathogen Escherichia coli O157:H7 in spinach. J. Food Prot 72, 1521–1530. [DOI] [PubMed] [Google Scholar]
- Moore MD, Goulter RM, Jaykus LA, 2015. Human norovirus as a foodborne pathogen: challenges and developments. Annu. Rev. Food Sci. Technol 6 (6), 411–433. [DOI] [PubMed] [Google Scholar]
- Park CH, Baskar TB, Park SY, Kim SJ, Arasu MV, Al-Dhabi NA, Kim JK, Park SU, 2016. Metabolic Profiling and Antioxidant Assay of Metabolites from Three Radish Cultivars (Raphanus sativus). Molecules 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu SH, Beaulieu JC, Prinyawiwatkul W, Ge BL, 2009. Effects of plant maturity and growth media bacterial inoculum level on the surface contamination and internalization of Escherichia coli O157:H7 in growing spinach leaves. J. Food Prot 72, 2313–2320. [DOI] [PubMed] [Google Scholar]
- Richards GP, 2012. Critical review of norovirus surrogates in food safety research: rationale for considering volunteer studies. Food Environ. Virology 4, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robilotti E, Deresinski S, Pinsky BA, 2015. Norovirus. Clin. Microbiol. Rev 28, 134–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM, 2011. Foodborne illness acquired in the United States-–major pathogens. Emerg. Infect. Dis 17, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Ingram DT, Patel JR, Millner PD, Wang X, Hull AE, Donnenberg MS, 2009. A novel approach to investigate the uptake and internalization of Escherichia coli O157:H7 in spinach cultivated in soil and hydroponic medium. J. Food Prot 72, 1513–1520. [DOI] [PubMed] [Google Scholar]
- Siddiq M, Roidoung S, Sogi DS, Dolan KD, 2013. Total phenolics, antioxidant properties and quality of fresh-cut onions (Allium cepa L.) treated with mild-heat. Food Chem. 136, 803–806. [DOI] [PubMed] [Google Scholar]
- Suzutani T, Ogasawara M, Yoshida I, Azuma M, Knox YM, 2003. Anti-herpesvirus activity of an extract of Ribes nigrum L. Phytother Res. 17, 609–613. [DOI] [PubMed] [Google Scholar]
- Warriner K, Ibrahim F, Dickinson M, Wright C, Waites WM, 2003. Internalization of human pathogens within growing salad vegetables. Biotechnol. Genet. Eng. Rev 20 (20), 117–134. [DOI] [PubMed] [Google Scholar]
- Warriner K, Namvar A, 2010. The tricks learnt by human enteric pathogens from phytopathogens to persist within the plant environment. Curr. Opin. Biotechnol 21, 131–136. [DOI] [PubMed] [Google Scholar]
- Wei J, Jin Y, Sims T, Kniel KE, 2010. Manure- and biosolids-resident murine norovirus 1 attachment to and internalization by Romaine lettuce. Appl. Environ. Microbiol 76, 578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Jin Y, Sims T, Kniel KE, 2011. Internalization of murine norovirus 1 by Lactuca sativa during irrigation. Appl. Environ. Microbiol 77, 2508–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]