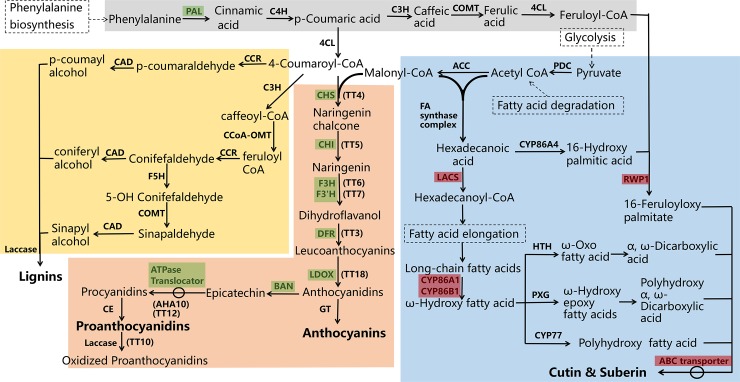
Fig 8. A simplified scheme of phenylpropanoid, lignin, flavonoid and cutin and suberin biosynthetic pathway.
These four pathways are shown with four different background colors. The down-regulated genes are marked as green and the up-regulated genes are marked as red. The other related biosynthetic pathways are placed in dotted boxes. PAL = phenylalanine ammonia lyase, C4H = Cinnamate-4-hydroxylase, C3H = Coumarate 3-hydroxylase, 4CL = 4-coumarate:CoA ligase, CCR = Cinnamoyl CoA reductase, CAD = Cinnamyl alcohol dehydrogenase, CCoA-OMT = Caffeoyl CoA O-methyltransferase, F5H = Ferulate 5-hydroxylase, COMT = Caffeic acid/5-hydroxyferulic acid O-methyltransferase, CHS = Chalcone synthase, CHI = Chalcone isomerase, F3H = Flavanone-3-hydroxylase, F3'H = Flavanone-3'-hydroxylase, DFR = Dihydroflavonol 4-reductase, LDOX = Leucoanthocyanidin dioxygenase, BAN = BANYULS, GT = Glucosyltransferase, CE = Condensing enzyme, AHA10 = Autoinhibited H(+)-ATPase isoform 10, TT = Transparent testa, ACC = acetyl-CoA carboxylase, PDC = Pyruvate dehydrogenase kinase, FA = Fatty acids, LACS = Long-chain acyl-CoA synthetase, CYP = Cytochrome P450 family, RWP1 = Reduced levels of wall-bound phenolics 1, HTH = fatty acid omega-hydroxy dehydrogenase, PXG = peroxygenase, ABC = ATP-binding cassette family.