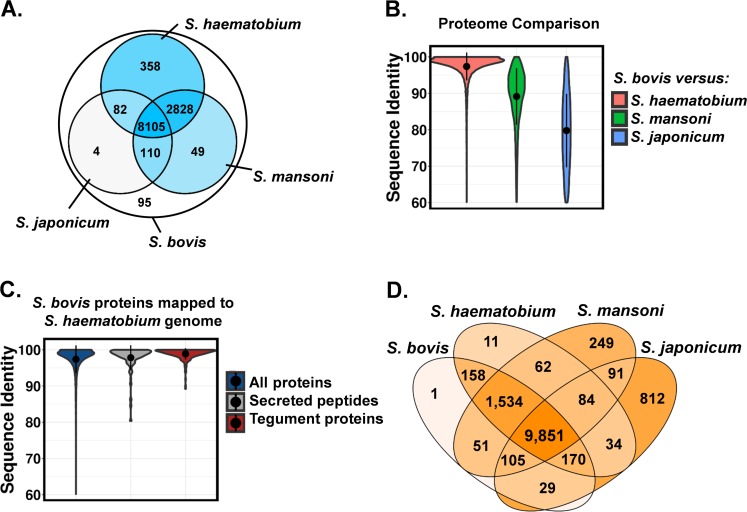
Fig 1. Comparison of the S. bovis proteome with other schistosome species.
A) S. bovis centred Venn diagram. The outer ring represents the 11,631 proteins predicted from the S. bovis genome, while the inner 3-way Venn diagram shows the subset of predicted S. bovis proteins that were present in the genomes of S. haematobium (Egypt isolate), S. mansoni and S. japonicum, respectively, based on protein sequence homology. The colours of the inner 3-way Venn diagram correspond to the number of proteins in each intersection from low (light grey) to high (dark blue). A Schistosoma core-proteome of 8,105 proteins was identified as those proteins that had sequence homology matches across all four species. 95 predicted proteins were unique for S. bovis. B) Sequence identity of S. bovis proteins and orthologous proteins from other schistosome species. Proteins of S. bovis and S. haematobium were significantly more conserved (p<10−15, two-tailed t-test) than the proteomes of S. bovis-S.-mansoni and S. bovis-S. japonicum. C) Sequence conservation of S. bovis tegument proteins and secreted peptides. S. bovis proteins were mapped to the S. haematobium genome sequence using Exonerate. Tegument proteins and secreted peptides showed an average sequence identity of 98.9% and 97.8%, respectively. All tegument proteins and secreted peptides included in this analysis were shared by both species. D) Analysis of the Schistosoma pan-genome. Proteomes of four Schistosoma species were concatenated, clustered into orthologous groups and mapped to each Schistosoma genome using Exonerate. Colours depict the number of proteins in each intersection from low (light orange) to high (dark orange). A pan-genome of 13,297 orthologous groups and a Schistosoma core-genome of 9,851 proteins were identified.