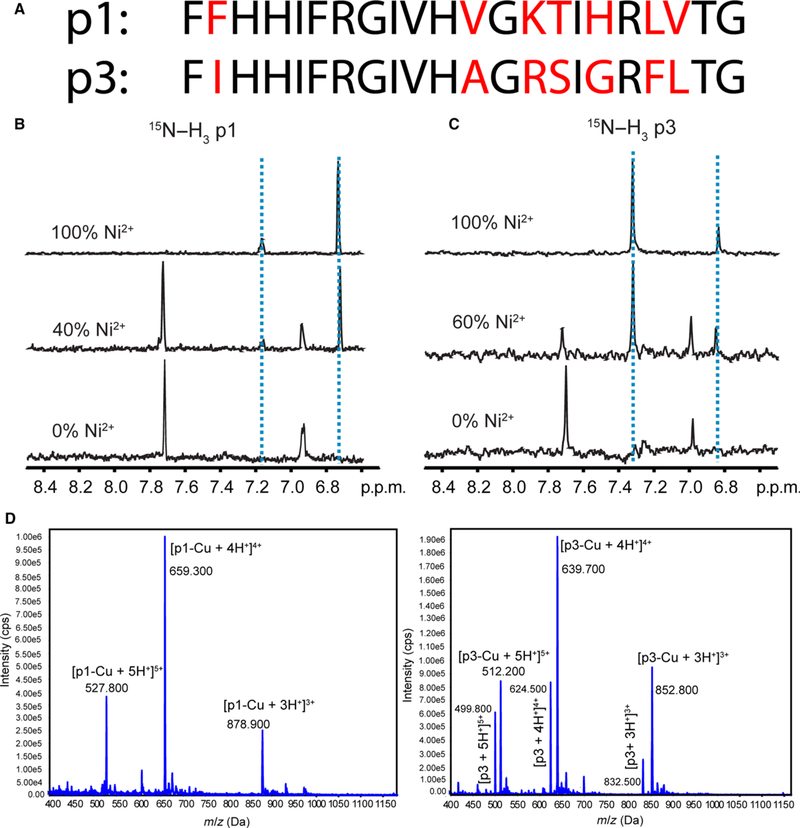
Fig. 1.
Structural characterization of copper binding to p1 and p3. (A) Amino acid sequences of p1 and p3 [19]. The nonconserved residues are shown in red. (B) and (C) One dimensional slices from solution NMR heteronuclear multiple quantum correlation (HMQC) spectra of 15N[α, δ, ε]-His3-labeled p1 (B) and 15N[α, δ, ε]-His3-labeled p3 (C) titrated with Ni2+ indicating perturbation of chemical shifts of d-proton (7.0 p.p.m.) and δ-proton (7.7 p.p.m.) of His3. (D) Mass spectrometry of piscidin exposed to Cu2+. Cu2+ and piscidin were mixed in a 1 : 1.2 molar ratio and subsequently injected into an electrospray ionization MS. Peaks corresponding to a 1 : 1 stoichiometry of binding appear in the mass spectra. Peaks at m/z 499.8, 624.5, and 832.5 in the bottom panel correspond to the +5, +4, and +3 charged states of naive p3.