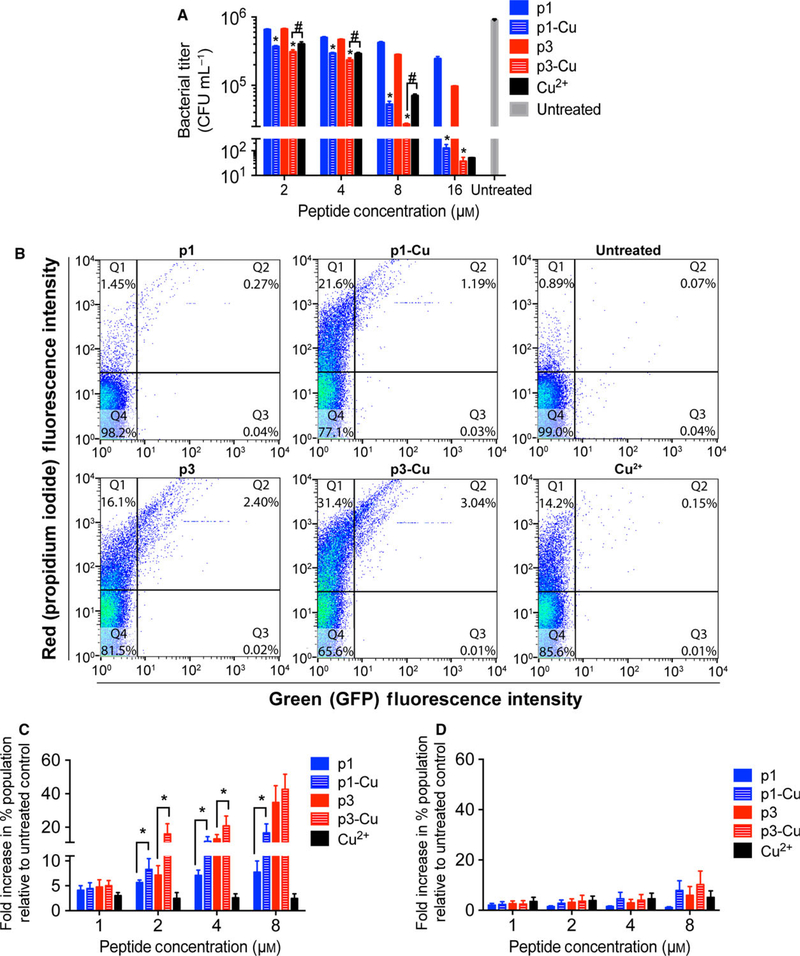
Fig. 7.
Activity of p1 and p3 against persister cells. (A) P. aeruginosa PA01 persisters were treated with the indicated concentrations of p1 and p3 for 3 h prior to CFU enumeration. Bars represent mean ± SEM (n = 3 triplicates). *P < 0.01, #P < 0.05 compared to the corresponding nonmetallated peptide. (B) Representative scatter plots from flow cytometric analysis of E. coli AT15 persisters treated with the indicated piscidin species and stained with PI. (C) Quantitative analysis of the increase in doubly fluorescent – GFP(+), PI(+) – cells (dead cells bearing DNA damage) following E. coli AT15 persister cell treatment with the indicated peptide concentrations. Bars represent mean ± SEM (n = 3). *P < 0.05. (D) Analysis of DNA damage in E. coli AT15 persister cells shows that number of GFP(−), propidium iodide (+) cells (dead cells without a damaged DNA; Q1 in scatter plots in (B)) is unchanged upon addition of copper.