Figure 1.
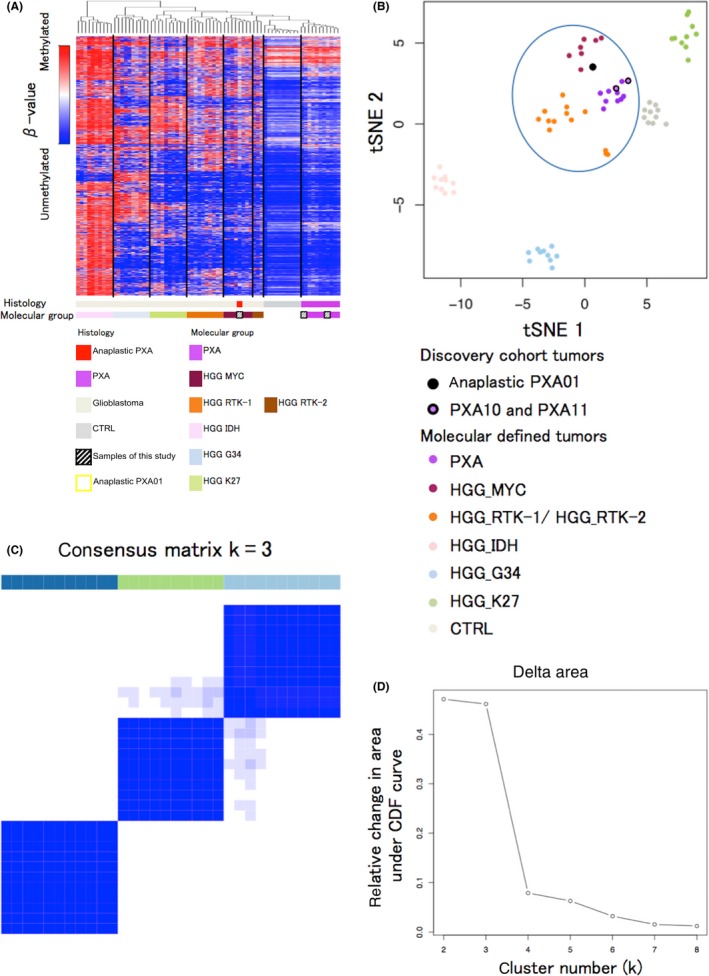
Genome‐wide methylation analysis. A, Unsupervised hierarchical clustering of DNA methylation and heatmap for the full tumor cohort based on the most variable 6152 methylated CpG island probes (standard deviation [SD] > 0.27) across the brain tumors methylation data sets. The probes are located within the CpG islands, whereas those on sex chromosomes are excluded. Anaplastic PXA01 was classified into the HGG_MYC type. B, A t statistic‐based stochastic neighbor‐embedding (tSNE) projection of the 450k methylation data. Anaplastic PXA01 and three clusters of GBM, ie, HGG_MYC, HGG_RTK (HGG_RTK‐1 and HGG_RTK‐2), and PXA clustered closely together (indicated by an oval). C, Heatmap for consensus matrix (k = 3) for tumors of anaplastic PXA01 and PXA with pediatric GBM of HGG_MYC and HGG_RTK (HGG_RTK‐1/‐2) types shows three distinct clusters, which is provided for the purpose of inner validation to confirm those relatively approximate classifiers in (B). These clusters are concordant with the unsupervised hierarchical clustering. D, Change in delta k for consensus matrix cumulative distribution function as k varies from 2 to 8, which showed the reliability of three distinct clusters. HGG, high‐grade glioma; PXA, pleomorphic xanthoastrocytoma
