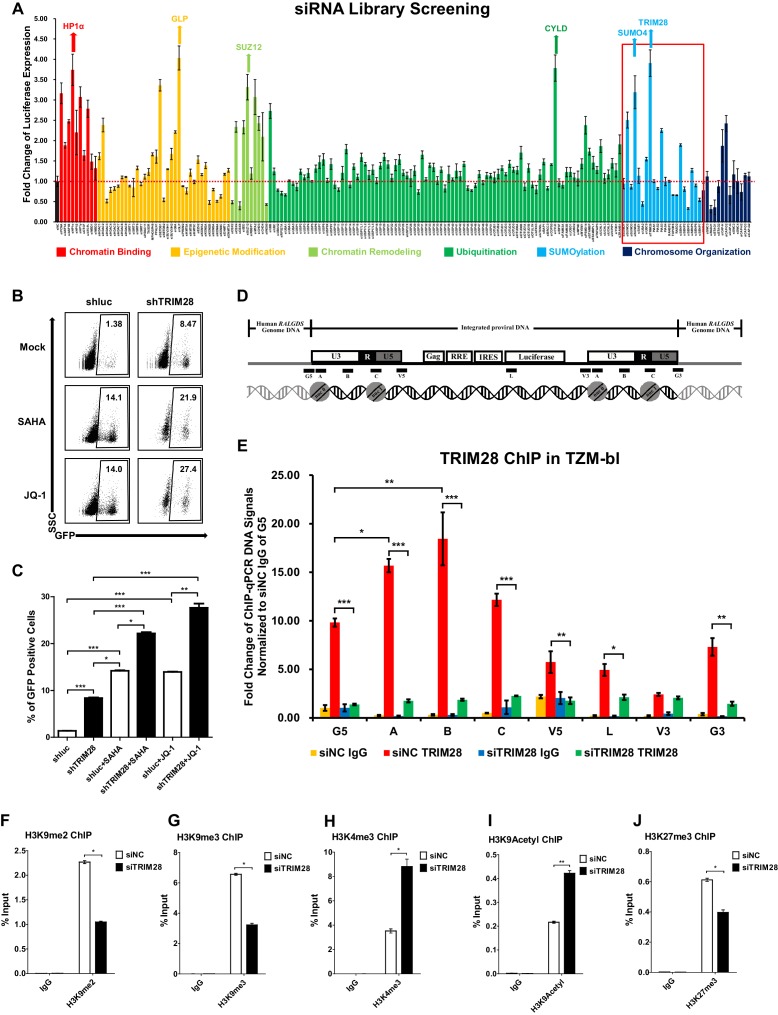
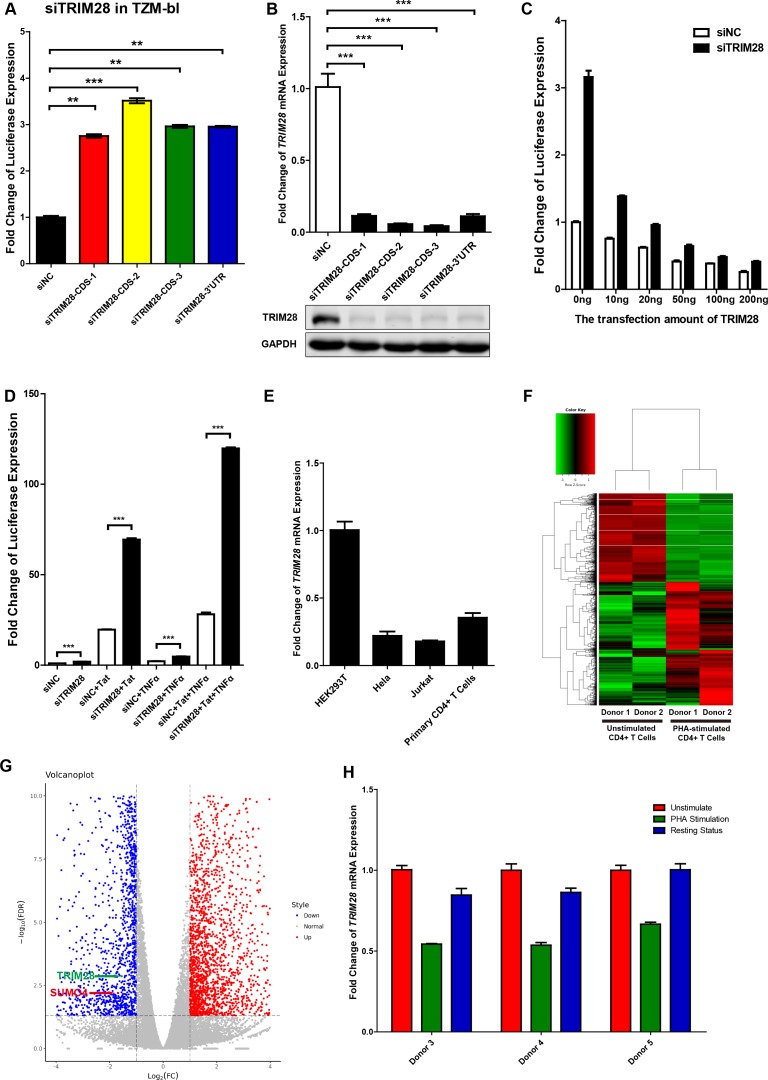
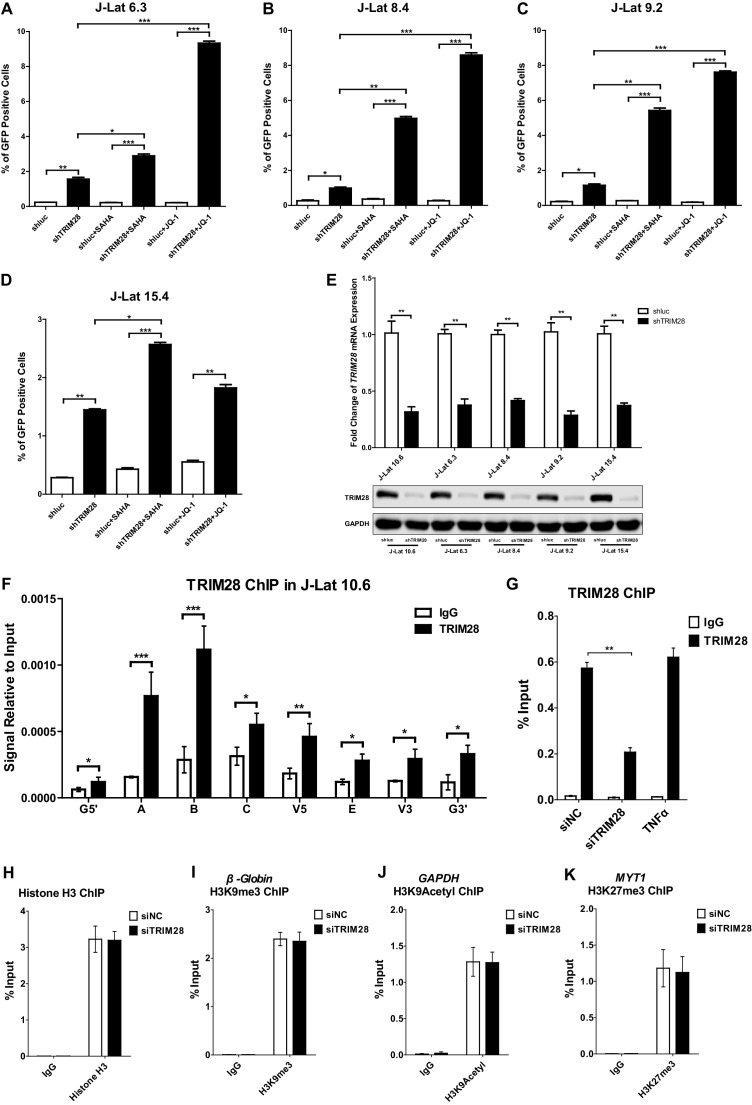
Figure 1. TRIM28 suppresses HIV-1 expression and contributes to HIV-1 latency.
(A) A siRNA library targeting 182 human genes was transfected into TZM-bl cell line, respectively. Three distinct siRNAs targeting each gene were transfected as a mixture. Forty-eight hours post-transfection, cells were harvested and the activity of luciferase from cell lysates was measured. Fold changes were calculated for each gene compared to negative control siRNA (siNC). (B–C) shRNA constructs were packaged into recombinant lentiviruses and infected J-Lat 10.6. The reactivation efficiency was measured by the GFP-positive percentage which was shown in the top right corner. SAHA and JQ-1 were used as positive controls. (D) Eight ChIP-qPCR primers targeting HIV-1 reporter provirus were designed. G5: Cellular DNA and viral 5’LTR junction; A: Nucleosome 0 assembly site; B: Nucleosome-free region; C: Nucleosome one assembly site; V5: Viral 5’LTR and gag leader sequence junction; L: Luciferase region; V3: Viral poly purine tract and 3’LTR junction; G3: Viral 3’LTR and cellular DNA junction. (E) ChIP assay with antibody against TRIM28 was performed in TZM-bl cell line. All the ChIP-qPCR DNA signals were normalized to siNC IgG of G5. (F–J) ChIP assays with antibodies against H3K9me2, H3K9me3, H3K4me3, H3K9Acetyl and H3K27me3 were performed in TZM-bl cell lines. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01, ***p<0.001.