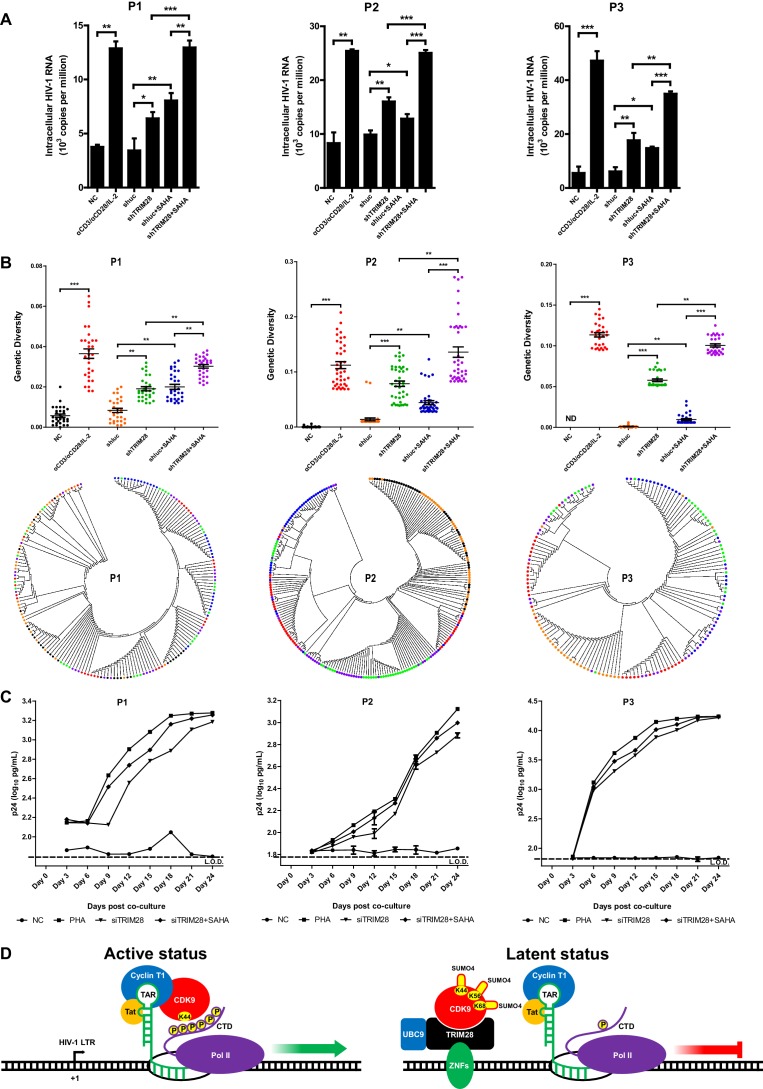
Figure 10. TRIM28 depletion reactivates latent HIV-1 in cells from HIV-1-infected individuals.
(A) shRNAs targeting luciferase and TRIM28 were packaged into lentiviruses and infected CD4+ T cells from HIV-1-infected individuals. Unstimulated CD4 +T cells were used as negative control (NC). Stimulation with αCD3/αCD28/IL-2 was used as positive control. Intracellular HIV-1 RNA was isolated and quantitated by qPCR. Experiments were conducted in three HIV-1-infected individuals. (B) The experiment setting was as in (A). Envelope V1 to V3 region from intracellular HIV-1 RNAs was reverse-transcribed and PCR-amplified. The PCR products were TA-ligated in pMD-18 T vector. At least 60 single clones were picked from each group and sequenced. The sequences from each group were aligned and the genetic diversity index was calculated and analyzed by Mann-Whitney U-test. The upper panel showed the statistical analysis results. The lower panel indicated the bootstrap consensus trees which were generated based on HIV-1 sequences. *p<0.05, **p<0.01, ***p<0.001. (C) Resting CD4+ T cells from HIV-1-infected individuals were isolated and nucleofected with siRNAs targeting negative control or TRIM28. Seventy-two hours later, PHA-stimulated uninfected CD4+ T cells were added into each group and co-cultured for another 27 days. The supernatants were collected and half-changed every 3 days. P24 antigens in supernatants were measured with ELISA and plotted in log10 scale. Dashed lines indicated the limit of detection (L.O.D.) of 50 pg/ml. Triplicates were represented by mean ±SEM. (D) Schematic of TRIM28-mediated HIV-1 latency.