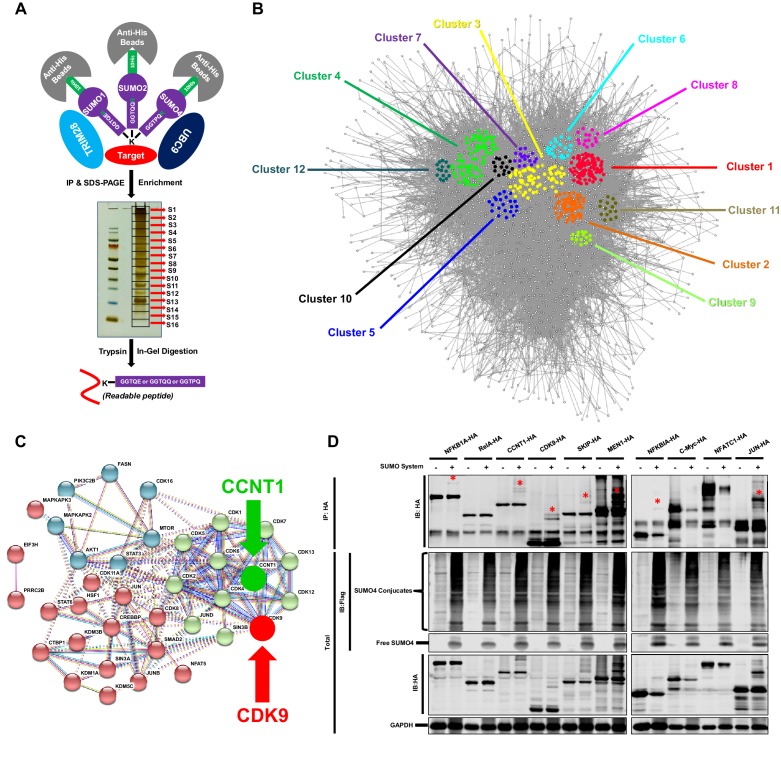
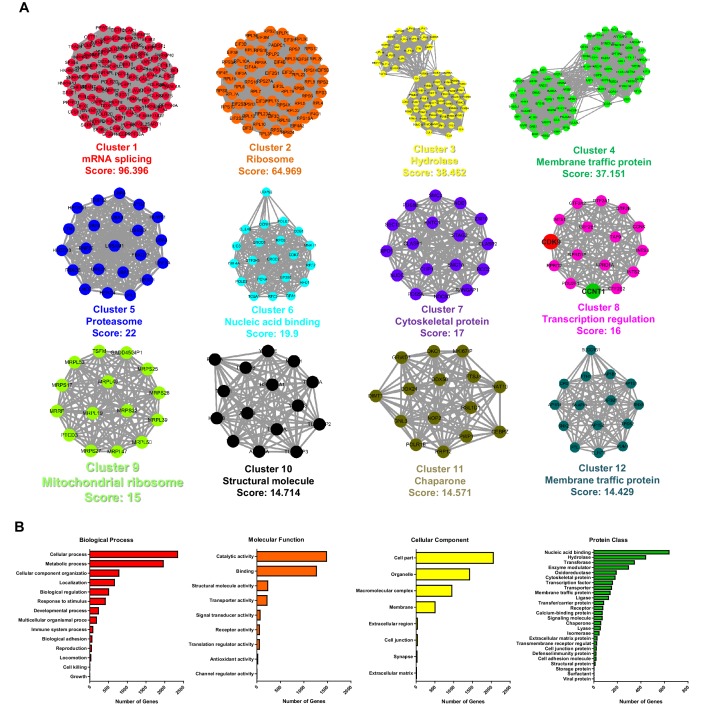
Figure 3. TRIM28 SUMOylates many transcription factors and transferases.
(A) Schematic of global site-specific SUMO-MS. His-tagged SUMO mutants were co-overexpressed with UBC9 and TRIM28. The SUMOylated proteins were enriched by His-tag beads and separated by SDS-PAGE. Gel fragments were excised and subjected to separate in-gel digestions. The digested peptides were desalted and analyzed by nanoscale LC-MS/MS. (B) SUMOylated proteins were analyzed with STRING. The network were further analyzed by MCODE. Twelve highly interconnected functional subclusters were extracted and shown in different colors. (C) Transferases and transcription factors were clustered by k-means clustering and visualized with STRING analysis. (D) Ten HA-tagged various transcriptional factors were overexpressed with Flag-tagged SUMO proteins, UBC9 and TRIM28. The targeted proteins were immunoprecipitated (IP) by anti-HA-tag beads followed by immunoblotting (IB) with anti-HA and –Flag antibodies. Asterisk (*) indicated the SUMOylated bands.