Figure 1.
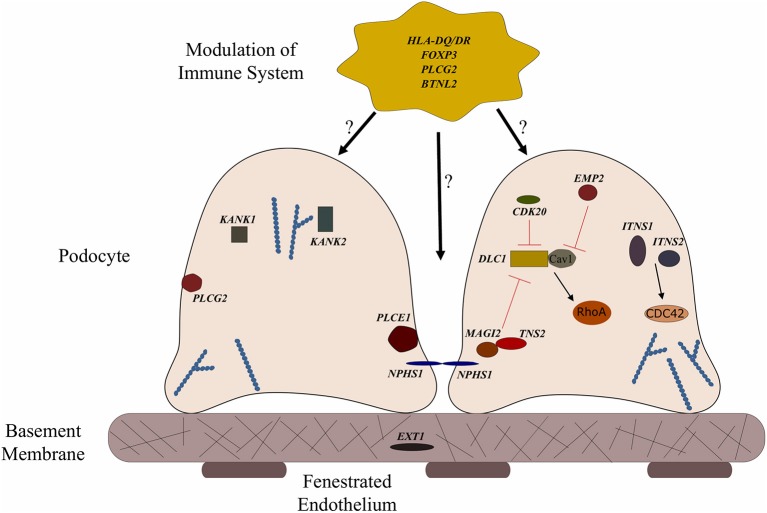
Genes associated with steroid sensitive nephrotic syndrome (SSNS). In the relatively rare cases of monogenic SSNS, the vast majority of genes that have been implicated in SSNS pathology locate to the glomerular filtration barrier and more specifically to the podocytes and slit diaphragm (Table 1). Among them are a key slit diaphragm protein Nephrin encoded by NPHS1 which is known to interact with MAGI2 and TNS2. These two proteins also interact as binding partners and along with CDK20 act to influence podocyte cytoskeletal activity through negative regulation of DLC1 activity (Red lines). DLC1 encodes s a RhoGTPase activating protein for RhoA and its activity can be modulated by the product of EMP2 through inhibition of the DLC1 binding partner Caveolin1 (CAV1). Intersectins 1 and 2 encoded by ITSN1 and ITSN2 act as GEFs for another small RhoGTPase, CDC42. The molecules encoded by KANK1 and KANK2 are also involved in RhoGTPase regulation and are known to affect actin polymerization. PLCE1 and PLCG2 encode proteins that modulate actin cytoskeleton and signaling at the slit diaphragm through calcium regulation. In addition to podocyte expression, PLCG2 is expressed in lymphocytes and it is known to affect immune response signaling. Evidence suggests that most cases of SSNS involve modulation of the immune system, though it is not clear exactly how this immune dysregulation affects the integrity of the glomerular filtration barrier. MHC Class II variants in the HLA-DR/DQ region are likely key components of this altered immune response. Other candidate loci include another MHC class II associated molecule BTNL2, as well as a regulator of T cell activity encoded by FOXP3. EXT1 is expressed in the glomerular basement membrane where it is involved in heparan sulfate biosynthesis.