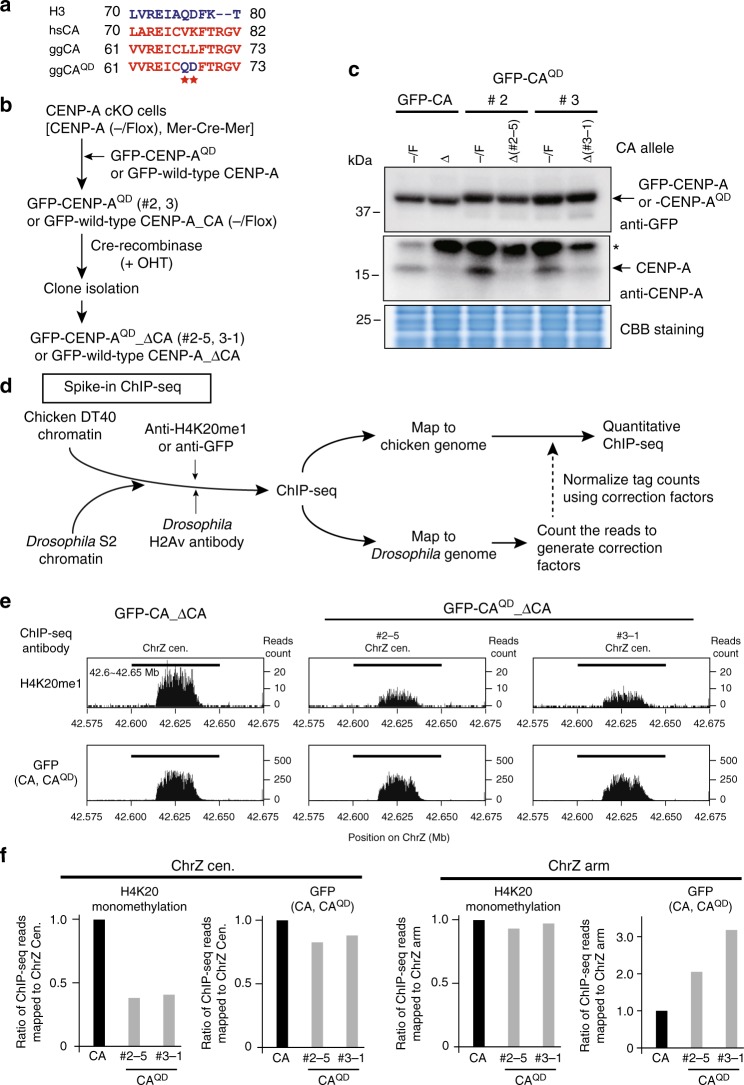
Fig. 4.
CENP-AQD mutation causes reduced H4K20 monomethylation in the DT40 centromere. a Alignment of the amino acid sequences of human histone H3.1, human CENP-A (hsCA), chicken CENP-A (ggCA), and chicken CENP-AQD (ggCAQD). The amino acid residues corresponding to the human CENP-A V76 and K77 residues are the chicken CENP-A L67 and L68 residues, respectively. b Experimental design to create CENP-A-deficient cells expressing either GFP-tagged CENP-A or CENP-AQD. c Immunoblot analyses with anti-GFP (Upper panel) and anti-CENP-A (Middle panel) antibodies, using whole-cell protein extracts from cells expressing GFP-tagged CENP-A (GFP-CA) or CENP-AQD before (-/F) and after (Δ) the endogenous CENP-A knockout. Two-independent cell lines were used for cells expressing CENP-AQD [#2 and #3]. Endogenous CENP-A depletion was confirmed (Middle panel). The asterisk indicates non-specific bands. Coomassie Brilliant Blue (CBB) staining is shown as a loading control (Lower panel). d Schematic representation of the Spike-in ChIP-seq. e Normalized ChIP-seq profiles with anti-H4K20me1 (Upper panels) and anti-GFP (Lower panels) around the centromere region on chromosome Z (42.575–42.675 Mb) in CENP-A-deficient DT40 cell lines, expressing either GFP-CENP-A (Left side panels) or GFP-CENP-AQD (#2-5; Middle and #3-1; Right panels). The centromeric accumulation of H4K20me1 was reduced in the cells expressing GFP-CENP-AQD (Upper middle and right panels). f Ratio of normalized ChIP-seq reads mapped to the chromosome Z centromere (ChrZ cen.: 42.6–42.65 Mb, Left two graphs). Ratios of reads mapped to the non-centromeric chromosome Z arm are also shown (ChrZ arm, Right two graphs). The reads of H4K20me1 mapped to the centromeric region in the GFP-CENP-AQD #2-5 and #3-1 cell lines were reduced to 0.38 and 0.41, respectively, as compared to the reads in cells expressing GFP-CENP-A. The mapped reads to the arm region were similar in both cells expressing GFP-CENP-A and GFP-CENP-AQD