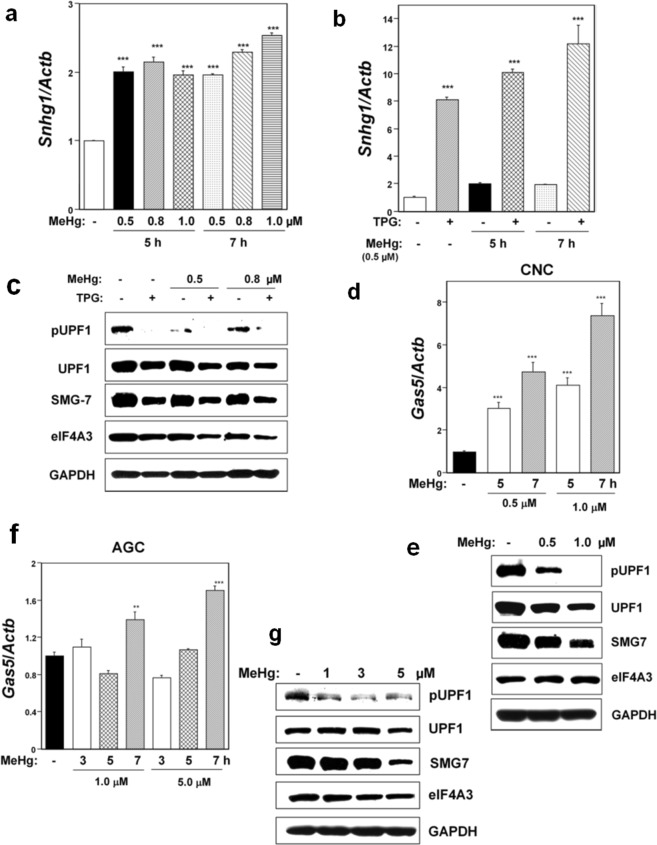
Figure 1.
Effects of MeHg or mild ER stress on NMD activity in C2C12-DMPK160 cells (a–c), CNCs (d,e), and AGCs (f,g). (a) RT-qPCR analysis of Snhg1 mRNA. The histogram depicts Snhg1 mRNA normalized to Actb mRNA presented as the fold-increase over non-pretreated controls. Values represent mean ± SE of three separate experiments. ***Significantly different from MeHg-untreated cells by one-way ANOVA followed by Bonferroni’s multiple comparison test (p < 0.001). (b) Effects of mild ER stress on NMD activity. C2C12-DMPK160 cells pretreated with TPG (0.2 μg/ml) for 16 h were exposed to 0.5 μM MeHg for 5 or 7 h. The histogram depicts Snhg1 mRNA normalized to Actb mRNA presented as the fold-increase over non-pretreated MeHg-untreated controls. Values represent mean ± SE (n = 3). ***Significantly different from TPG-untreated cells by one-way ANOVA followed by Bonferroni’s multiple comparison test (p < 0.001). (c) Western blotting analyses of NMD components’ protein expression. Total cell lysates prepared 7 h after exposure to 0.5 or 0.8 μM MeHg were analyzed using the indicated antibody probes. Cropped blots are shown; all gels were run under the same experimental conditions. Raw data are shown in Supplementary Fig. 1c with dividing lines and with quantitative data. (d,f) RT-qPCR analysis of Gas5 mRNA. The histogram depicts Gas5 mRNA normalized to Actb mRNA represented as the fold-increase over MeHg-untreated controls. Values shown are the mean ± SE of three separate experiments. **,***Significantly different from MeHg-untreated cells by one-way ANOVA followed by Bonferroni’s multiple comparison test (**p < 0.01, ***p < 0.001). (e,g) Western blotting analyses of expression of NMD component proteins. Total cell lysates prepared 24 h for CNCs or 9 h for AGCs after exposure to the indicated concentration of MeHg were analyzed using the indicated antibody probes. Cropped blots are shown; all gels were run under the same experimental conditions. Raw data are shown in Supplementary Fig. 1e, g with dividing lines and with quantitative data.