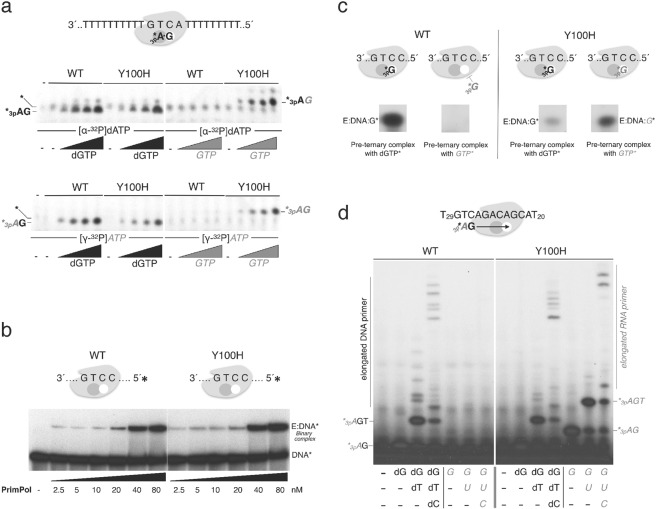
Figure 3.
Ribonucleotides are valid substrates for the Y100H variant during primer synthesis. (a) Scheme on the top shows PrimPol in complex with the GTCA template oligonucleotide and the two nucleotides forming the initial dimer. The autoradiograph shows dimer formation (primase activity) either by wild-type (WT) PrimPol or Y100H (400 nM) using [α-32P]dATP (upper panel) or [γ-32P]ATP (lower panel) as the 5′-site nucleotide (16 nM), and increasing concentrations of either dGTP or GTP as the incoming 3′-site nucleotide (0, 10, 50, 100 µM). (b) Binary complex formation, measured by EMSA, between WT PrimPol or Y100H and labeled 60-mer DNA template GTCC (1 nM), using the indicated PrimPol concentration (2.5, 5, 10, 20, 40 and 80 nM) (c) Pre-ternary complex formation measured by EMSA between WT PrimPol or Y100H (1 µM), 60-mer DNA template GTCC and either [α-32P]dGTP or [α-32P]GTP (16 nM). (d) DNA or RNA primers synthesized using as template 5′-T20ACGACAGACTGT29 -3′ to allow elongation beyond the dimer. Products were labeled with [γ-32P]ATP (16 nM) as the 5′nucleotide and each subsequent nucleotide (dGTP, dTTP, dCTP) was added (10 µM) as indicated in the figure. Full length gels corresponding to parts a to d are shown in Supplemental Fig. 6. The autoradiographs shown in this figure are representative of at least 3 independent experiments.