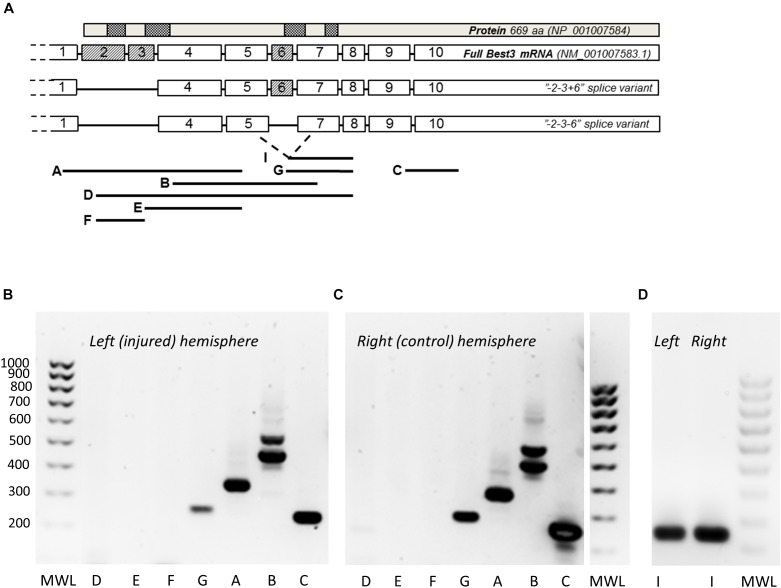
FIGURE 2.
PCR analysis of Best3 mRNA alternative splicing in mouse brain with and without HI injury. Alternative splicing of Best3 mRNA was studied by a set of primers (A) spanning the areas of expected splicing of exons 2, 3, and 6 in different combinations as described in detail previously (Golubinskaya et al., 2015). The results of these PCR experiments are presented in (B–D). Total Best3 mRNA can be detected by primer pair C localized in exons 9 and 10 in both healthy (C) and injured brain (B). Splice variants of Best3 with exons 2 and 3 present were not detected, neither in healthy brain nor in injury (pairs of primers: A, localized in exons 1 and 5; D in exons 2 and 7; E in exons 3 and 5; F in exons 2 and 3). Exon 6 can be either present or spliced out in Best3 mRNA. Primer pair B spanning exons 4–7 gives two bands, the heavier band corresponds to “+6” and the lighter one to “–6” splice variants. The pair of primers G localized in exons 6 and 8 shows a band corresponding to “+6” splice variant. Primer pair I detects the “–6” variant (D). Primer pairs G and I can potentially detect more than one transcript corresponding to “+6” and “–6” variants depending on possible splicing of exon 10. Positive controls for these primers have been published previously by our group (Golubinskaya et al., 2015). MWL, molecular weight ladder.