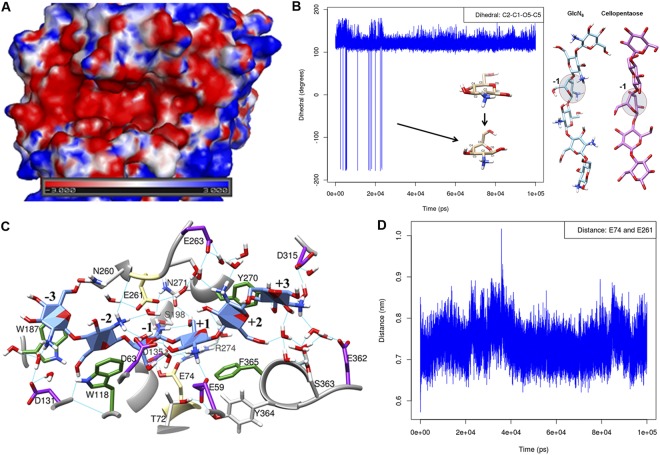
Figure 4.
Molecular modeling studies defining the property of the binding site. (A) Electrostatic surface potential map of CSN-MN representing the very acidic binding site; red, acidic regions; blue, basic regions. (B) The 4C1 chair conformation was observed as the major conformation during simulation, changes in the dihedral angles at the different time points of simulation at subsite (−1) indicate switches in the pyranose ring conformation (see also Fig. S5). (C) Enzyme-substrate interaction map between CSN-MN and GlcN6 as a substrate showed all possible interactions; catalytic residues E74 and E261 are highlighted in khaki color, aromatic residues making stacking interactions with the substrate are colored in green, and acidic residues are colored in violet. (D) The distance measured between the catalytic residues E74 and E261 during simulation (average 7.4 Å) indicates that CSN-MN is an inverting enzyme.