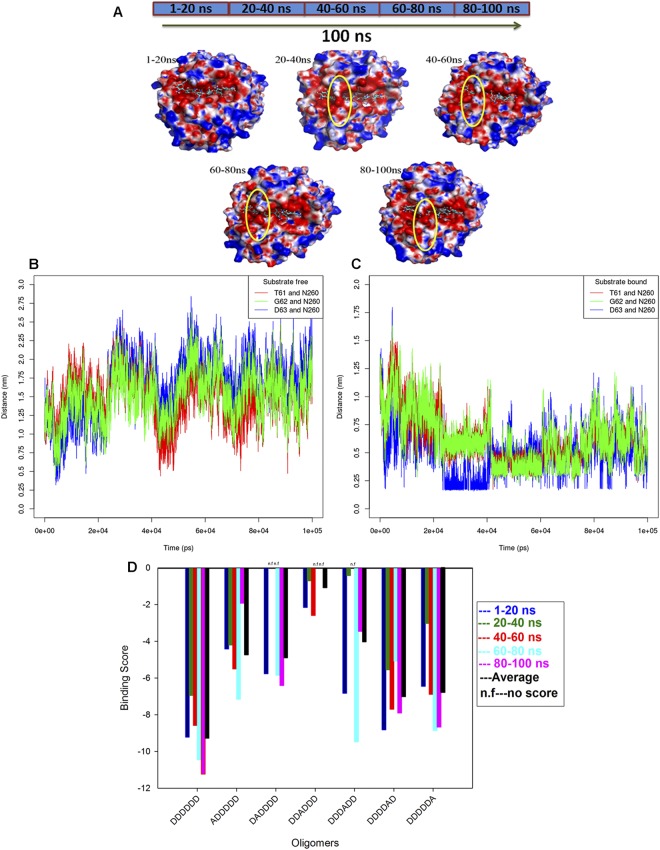
Figure 6.
Conformational changes occur at the binding site of the enzyme in the presence of substrate, and ensemble docking results. (A) Average structures of substrate-bound CSN-MN derived from clusters 1–20 ns, 20–40 ns, 40–60 ns, 60–80 ns, and 80–100 ns, displaying open and closed surfaces at the binding site of the enzyme in the presence of substrate. (B) Distance between the amino acid residues from loop L1 (T61, G62, D63) and loop L2 (N260) in substrate-free CSN-MN, showing that during simulation, residues from loops occasionally approached closely. (C) Distance between the amino acid residues from loop L1 (T61, G62, D63) and loop L2 (N260) in substrate-bound CSN-MN, showing that during simulation, residues from loops approached to come into the range of interaction and were involved in forming the closed surface at subsite (−2). (D) Binding score of fully deacetylated chitosan hexamer to CSN-MN, displaying a change in the score with a change in the conformation of enzyme or substrate, and calculated average scores. The highest score was measured for oligomer DDDDDD, and the lowest for DDADDD, indicating subsite (−1) specificity (see also Fig. S8).