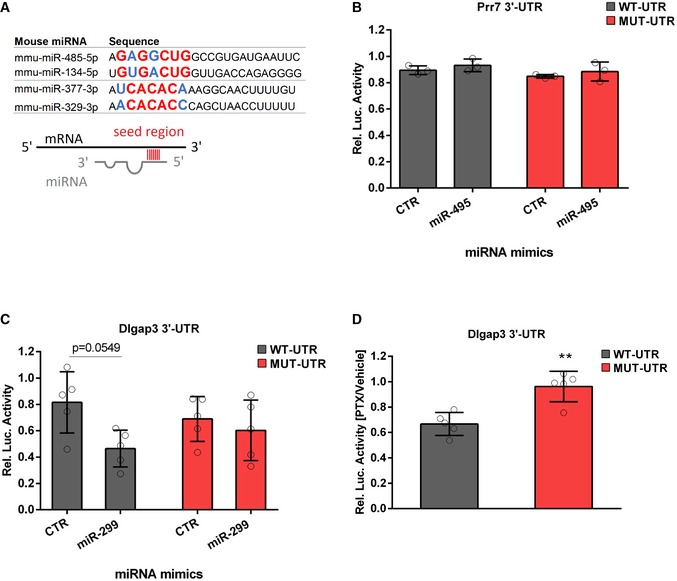
miRNAs of the miR379‐410 cluster sharing similar seed regions: miR485‐5p/miR‐134‐5p and miR‐377‐3p/miR‐329‐3p. Seed regions (nucleotide 2–8) are highlighted in bigger letters, conserved nucleotides in red and those that allow wobble base pairing in blue.
3′UTR luciferase reporter gene assays in cultured rat cortical neurons using the Prr7 3′UTR reporter gene (wt or seed mutant) together with miR‐495 or control mimic, respectively (n = 3 per group; wt: P = 0.7775; mut: P = 0.7797; mimic: F
1,8 = 1.816, P = 0.2147; utr‐construct: F
1,8 = 2.973, P = 0.1229; mimic × utr‐construct: F
1,8 = 9.47e‐006, P = 0.09976); two‐way ANOVA.
3′UTR luciferase reporter gene assays in cultured rat cortical neurons using the Dlgap3 3′UTR reporter gene (wt or seed mutant) together with miR‐299 or control mimic, respectively (n = 5 per group; wt: P = 0.0549; mut: P = 0.8982; mimic: F
1,16 = 6.145, P = 0.0247; utr‐construct: F
1,16 = 0.005108, P = 0.9434; mimic × utr‐construct: F
1,16 = 2.246, P = 0.1534); two‐way ANOVA.
3′UTR luciferase reporter gene assays in cultured rat hippocampal neurons using the Dlgap3 3′UTR reporter gene (wt or seed mutant) after 48 h PTX stimulation (n = 5 per group, t
8 = 4.397, **P = 0.0023); unpaired Student's t‐test.
Data information: Data are presented as mean ± s.d.