Figure 2. Indirect imaging of the degradative functions of ubiquitin.
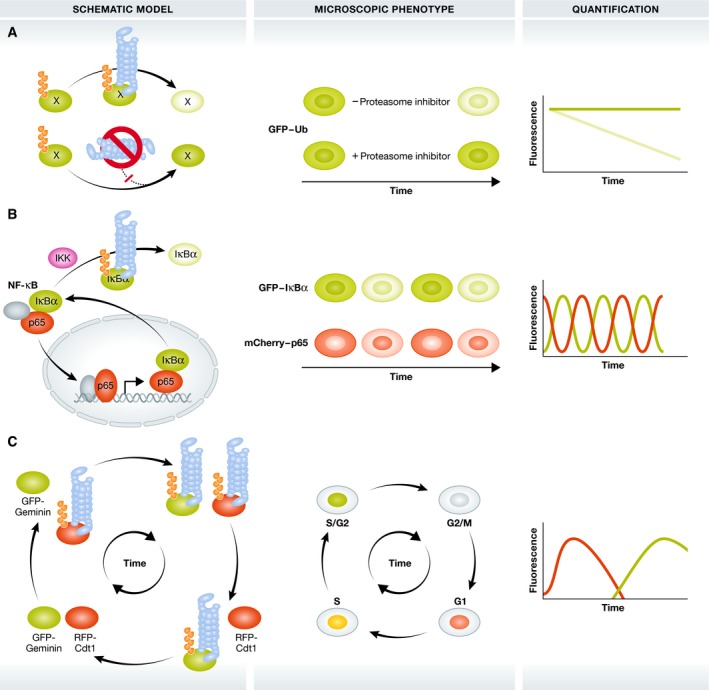
Approaches to image the proteasome‐related functions of Ub in the control of protein stability and breakdown. Darker and lighter green colours indicate the accumulation and breakdown of proteins, respectively. (A) GFP‐labelled Ub or model substrates are modified with degradative Ub signals and degraded by the 26S proteasome (blue barrel). Proteasome inhibition induces stabilization and accumulation of these GFP reporters as ubiquitinated forms. (B) Upon IKK activation, GFP‐tagged IκBα becomes modified with K48‐linked polyubiquitin chains and degraded by the 26S proteasome. This releases mCherry‐RelA/p65 that subsequently translocates in the nucleus to control NF‐κB‐dependent gene expression. IĸBα is among these NF‐κB target genes and shuttles back into the cytosol, creating dynamic NF‐κB degradation/translocation loops. Grey ovals: additional NF‐κB transcription factors (C) FUCCI: co‐expression of the UPS substrates GFP‐Geminin and RFP‐Cdt1 allows microscopic analysis of cell cycle phases by phase‐dependent Ub‐dependent degradation of GFP‐Geminin and RFP‐Cdt1.
