Fig. 1.
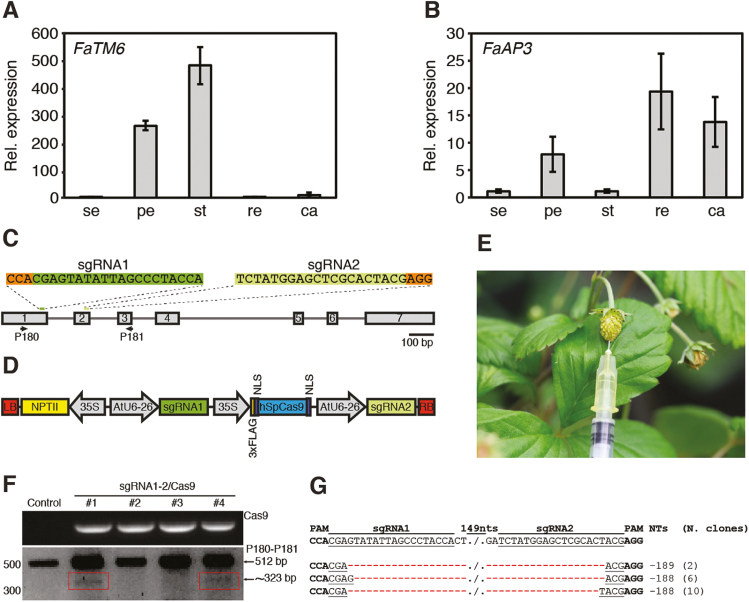
Expression of AP3 and TM6 genes in the cultivated strawberry (Fragaria × ananassa), construct design, and evaluation of CRISPR-Cas9-based editing of the wild strawberry, Fragaria vesca. Relative expression of the F. × ananassa TM6 gene (FaTM6) (A) and AP3 gene (FaAP3) (B) in sepals (se), petals (pe), stamens (st), receptacles (re), and carpels (ca) of F. × ananassa flowers at stage 12 as determined by qRT-PCR. Data are means (±SD) of three biological replicates with three technical replicates each. (C) The F. vesca TM6 (FveTM6) locus, including exons (boxes) and introns (lines). sgRNAs 1 and 2 are represented in green and PAM sequences in orange. The primers used for characterization of CRISPR/Cas9 editing are indicated. (D) Schematic representation of the two sgRNAs and Cas9 expression cassettes in a single binary vector pCMABIA2300 (sgRNA1-2/Cas9 vector). (E) Green fruit agro-infiltrated with the sgRNA1-2/Cas9 vector. (F) PCR to detect indels in FveTM6. Top panel: PCR of Cas9 in four fruits that transiently expressed the vector. Bottom panel: PCR with P180 and P181 (Supplementary Table S2) showing a ~323-bp band (red boxes) in fruits #1 and #4 in addition to the wild-type band (512 bp). (G) Alignment of sequences obtained after the purification, cloning, and Sanger sequencing of the ~323 bp band from fruits #1 and #4. Fragments with 189- and 188-bp deletions resulting from simultaneous DSBs in both target sites were detected in two and 16 clones, respectively.