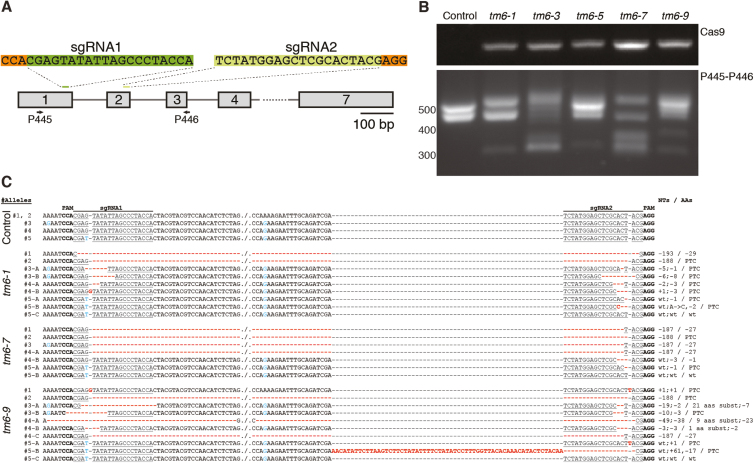
Fig. 2.
Identification of CRISPR/Cas9-induced mutations in the Fragaria × ananassa TM6 allele (FaTM6). (A) Schematic representation of the positions of the sgRNAs in the F. vesca TM6 (FveTM6) locus. The primers used for the analysis in agarose gels and for deep sequencing are represented (P445 and P446; Supplementary Table S2). (B) Top panel: identification of the Cas9 gene in transgenic tm6 lines. Bottom panel: detection of mutations at the FaTM6 locus using the P445 and P446 primers. (C) Sequence alignment obtained by high-throughput amplicon sequencing in control and tm6 mutant lines. PAM sequences are marked in bold; sgRNAs are underlined; blue indicates distinctive SNPs among FaTM6 alleles; bold red indicates mutations induced by CRISPR/Cas9-mediated editing. PTC, premature termination codon.