Figure 1.
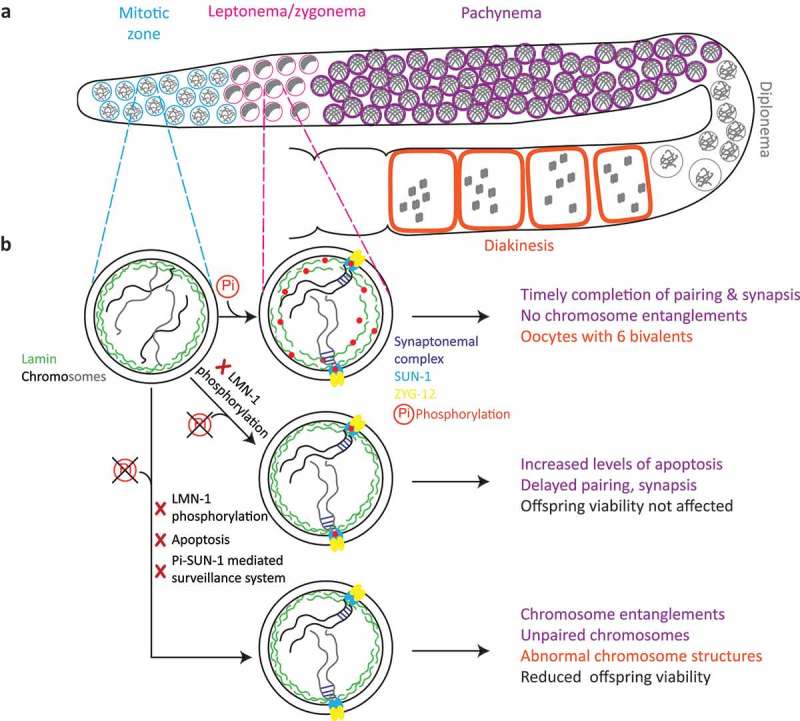
Impact of LMN-1 phosphorylation during meiosis in the C. elegans germline: (a) Schematic representation of meiotic progression along the C. elegans germline. Mitotic nuclei (blue) enter meiosis (pink), where the chromatin acquires a polarized configuration as a consequence of chromatin reorganization and chromosome movement. Nuclei then enter pachynema (purple) and homologous chromosomes are aligned in a side-by-side orientation. At diplonema (gray), chromatin condenses; during diakinesis (orange), oocytes with six bivalents can be seen in the wild type. (b) In interphase nuclei in the mitotic zone, non-phosphorylated lamin forms a rigid network. Upon meiotic entry, the lamina network is phosphorylated and adopts a more open structure. In leptonema/zygonema, chromosomes move, homologous chromosomes align, followed by assembly of the synaptonemal complex (blue lines) between homologs. Inhibition of phosphorylation renders the lamina network less detergent-soluble upon meiotic entry, which slows chromosome movement and delays pairing and synapsis. However, nuclei with abnormal chromosomes are culled by apoptosis during pachynema and overall offspring viability of the phospho-mutant is not affected. Deactivation of the apoptotic machinery or the phospho-SUN-1-mediated surveillance system combined with a more rigid lamina network leads to pachytene nuclei with chromosome entanglements and interlocks. Therefore, the resultant diakinesis oocytes have chromosomes with abnormal structures, which reduces offspring viability.
