Fig. 1.
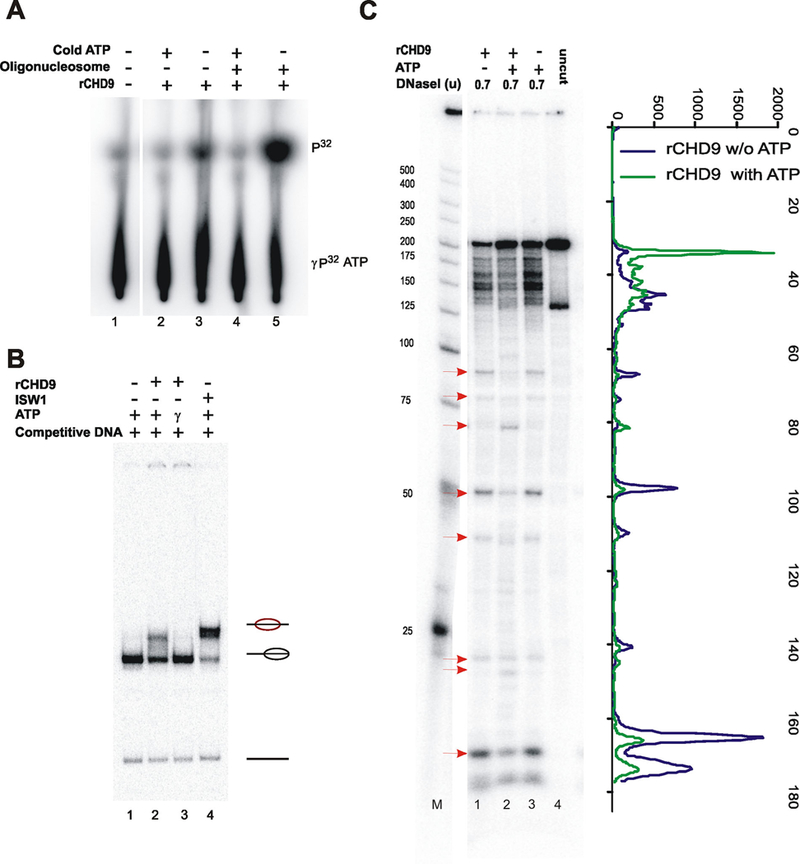
rCHD9 is a nucleosome stimulated, ATP-dependent chromatin remodeling protein. A: rCHD9 in the presence of (γ−32P)ATP and in the presence or absence of oligonucleosomes. Reaction products were separated on PEI-cellulose. B: Mononucleosomes (radiolabeled 601 fragment) were incubated with rCHD9 with ATP or non-hydrolyzable ATP-γ-S and nucleosome mobilization was monitored. Reactions were separated on a native 5% polyacrylamide gel, dried and exposed on a phosphorimager. The positions of nucleosomes (and their relative position on the fragment) and the free DNA are shown to the side. ISWI is used as a positive control in the sliding assay. C: Reconstituted nucleosomes were treated (as in B) followed by DNaseI digestion, with the indicated amounts, for 5 min. Reaction products were separated on 6% polyacrylamide-urea gels. Active chromatin remodeling proteins alter the pattern of DNaseI cutting on positioned nucleosomes. Red arrows reflect sites of increased or decreased cutting as a result of ATP-dependent chromatin remodeling.
