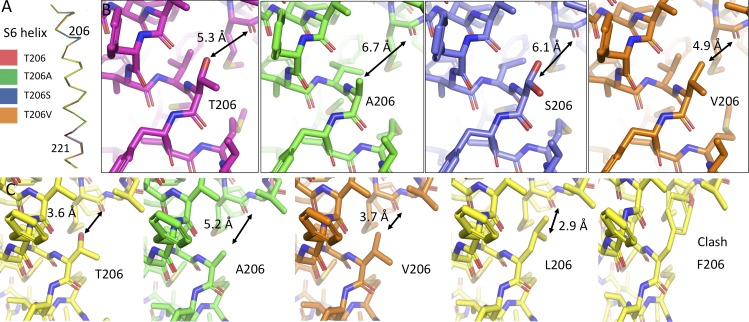
Figure 8.
The structures of Thr206 mutants. (A) Side view of the isolated S6 helices of NaVAbΔ28 (dark red), NaVAbΔ28/T206A (green), NaVAbΔ28/T206S (blue), and NaVAbΔ28/T206V (orange). (B) Stick models are shown for each of the mutations in A, with focus on the 206 position of the S6 helix and the nearby portion of the pore helix. Coloring is as in A. Distances between the centers of the atoms of the side chain at position 206 and the main-chain carbonyl of Met174 are displayed in black in order to illustrate the differences among these mutations. Two rotamers are shown for T206S (blue), as determined in the crystal structure. (C) Hypothetical positions of Thr206 mutations in the open state (PDB accession no. 5VB2), with (from left to right) Thr206 (WT), T206A, T206V, T206L, and T206F. In each case, the model was created by substituting the Thr206 position with the amino acid shown, without energy minimization or any other rotamer selection. Rotamers selected are the most commonly encountered in the PDB and are meant to be illustrative of space constraints.