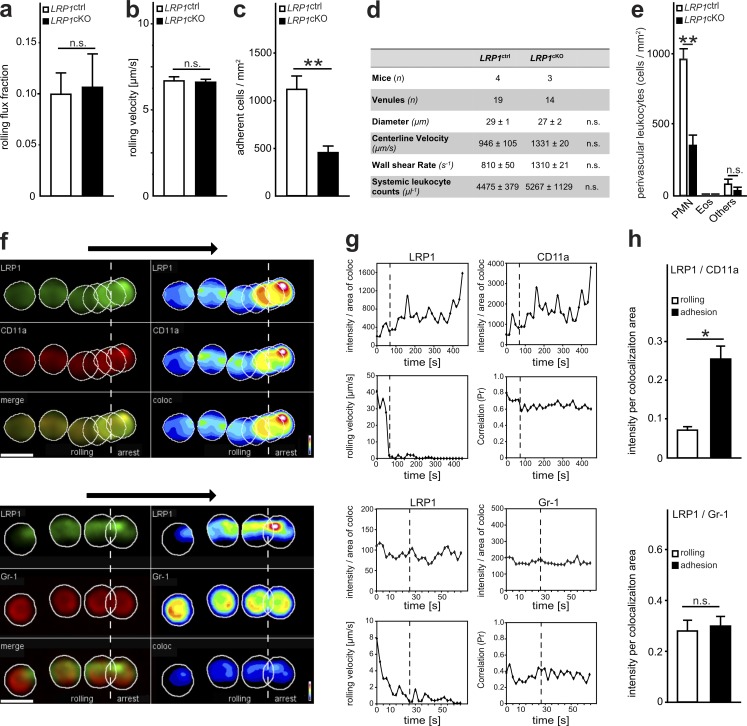
Figure 4.
The MK receptor LRP1 is critical for leukocyte recruitment. (a–d) Intravital microscopy of cremaster muscle postcapillary venules of LRP1ctrl and LRP1cKO mice 2 h after intrascrotal application of TNFα. n = 18 venules from four LRP1ctrl mice, n = 14 venules from three LRP1cKO mice. (a) Leukocyte rolling flux fraction (LRP1ctrl, 0.097 ± 0.020 [%]; LRP1cKO, 0.104 ± 0.032 [%]). (b) Leukocyte rolling velocity including 79 cells from four LRP1ctrl mice and 71 cells from three LRP1cKO mice (LRP1ctrl, 6.74 ± 0.24 µm/s; LRP1cKO, 6.66 ± 0.17 µm/s). (c) Number of adherent leukocytes per mm2 (LRP1ctrl, 1122.2 ± 133.2 cells/mm2; LRP1cKO, 456.0 ± 60.6 cells/mm2). (d) Hemodynamic parameters of investigated vessels. (e) Differential cell counts of perivascular leukocytes as analyzed histologically in cremaster muscle whole mounts after Giemsa staining 2 h after intrascrotal application of TNFα. Eos, eosinophils; Others, lymphocytes, basophils and monocytes; n = 47 venules from four LRP1ctrl mice, n = 28 from three LRP1cKO mice (PMN: LRP1ctrl, 958.3 ± 75.2 cells/mm2, LRP1cKO, 354.1 ± 65 cells/mm2). (f–h) Flow chamber assays with mPMN using spinning disk confocal microscopy after coating with P-selectin, ICAM-1, and CXCL1. (f) The intensity of LRP1 and CD11a as well as LRP1 and Gr-1 expression together with the merged (merge) or colocalized (coloc) intensity of one single representative cell is depicted during rolling and adhesion. Arrows indicate direction of flow. Right panels demonstrate the intensity of LRP1 and CD11a cell surface expression using pseudocolors (heat map). (g) Analysis of the intensity per area of colocalization of LRP1 or CD11a as well as of LRP1 or Gr-1, kinetics of rolling velocity and correlation of colocalization during rolling and adhesion of one representative cell. (h) Mean intensity per area of colocalization of LRP1 and CD11a (rolling, 0.071 ± 0.009 arbitrary units [a.u.]; adhesion, 0.255 ± 0.033 a.u.) as well as of LRP1 and Gr-1 (rolling, 0.280 ± 0.042 a.u.; adhesion, 0.299 ± 0.037 a.u.) during rolling and adhesion. LRP1/CD11a: n = 23 cells/five experiments, LRP1/Gr-1: n = 11 cells/three experiments. To determine P values, an unpaired Student's t test was performed for a–c and h, and ANOVA on ranks with Dunn’s multiple comparisons test was used for e. For all panels, *, P < 0.05; **, P < 0.01; n.s., not significant. Data are presented as mean ± SEM.