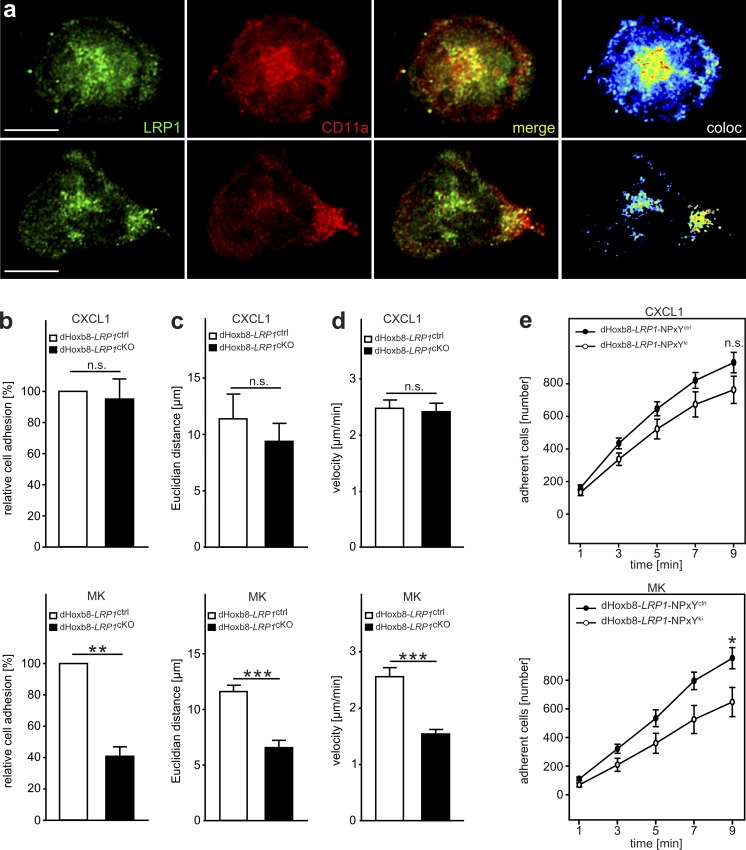
Figure 5.
LRP1-mediated PMN recruitment is MK dependent. (a) Representative images of dHoxb8-LRP1ctrl cells on microflow chambers coated with recombinant P-selectin, ICAM-1, and MK. Cells were perfused through chambers with a shear rate of 1 dyne/cm2. Staining of cells was conducted using specific anti-LRP1 and anti-CD11a Abs, and imaging was performed using STED nanoscopy. Images show expression of LRP1 (green), CD11a (red), merge (yellow), and colocalization (coloc; pseudocolors [heat map]) of LRP1 and CD11a. The top panel illustrates an adherent cell and the lower panel a migrating cell. (b) Induction of adhesion of dHoxb8-LRP1ctrl and dHoxb8-LRP1cKO cells under flow conditions (1 dyne/cm2) in microflow chambers coated with immobilized P-selectin, ICAM-1, and CXCL1 (dHoxb8-LRP1ctrl, 100.0 [%]; dHoxb8-LRP1cKO, 95.1 ± 12.7 [%]) or MK (dHoxb8-LRP1ctrl, 100.0 [%]; dHoxb8-LRP1cKO, 40.8 ± 6.0 [%]) as indicated. n = 4. (c and d) Migration of dHoxb8 cells under flow conditions. Euclidean distance (c; CXCL1: dHoxb8-LRP1ctrl, 11.4 ± 2.2 µm; dHoxb8-LRP1cKO, 9.4 ± 1.6 µm; MK: dHoxb8-LRP1ctrl, 11.6 ± 0.55 µm; dHoxb8-LRP1cKO, 6.56 ± 0.65 µm) as well as migration velocity (d; CXCL1: dHoxb8-LRP1ctrl, 2.47 ± 0.25 µm/min; dHoxb8-LRP1cKO, 2.41 ± 0.26 µm/min; MK: dHoxb8-LRP1ctrl, 2.56 ± 0.16 µm/min; dHoxb8-LRP1cKO, 1.54 ± 0.08 µm/min) of dHoxb8-LRP1ctrl (white bar) and dHoxb8-LRP1cKO (black bar) under flow conditions (1 dyne/cm2). Flow chambers were coated with P-selectin, ICAM-1, and CXCL1 or MK as indicated. dHoxb8-LRP1ctrl cells on CXCL1: n = 3 experiments/214 cells; dHoxb8-LRP1cKO cells on CXCL1: n = 3 experiments/303 cells. dHoxb8-LRP1ctrl cells on MK: n = 4 experiments/280 cells; dHoxb8-LRP1cKO cells on MK: n = 4 experiments/197 cells. (e) Induction of adhesion of dHoxb8-LRP1-NPxYctrl and dHoxb8-LRP1-NPxYki cells under flow conditions (1 dyne/cm2) in microflow chambers coated with immobilized P-selectin, ICAM-1, and CXCL1 (top diagram; dHoxb8-LRP1-NPxYctrl, 929.9 ± 62.9 cells; dHoxb8-LRP1-NPxYki, 762.8 ± 83.7 cells) or MK (bottom diagram; dHoxb8-LRP1-NPxYctrl, 953.3 ± 74.2 cells; dHoxb8-LRP1-NPxYki, 647.3 ± 101.8 cells). n = 6. To determine P values, an unpaired Student's t test was performed. For all panels, *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. Data are presented as mean ± SEM.