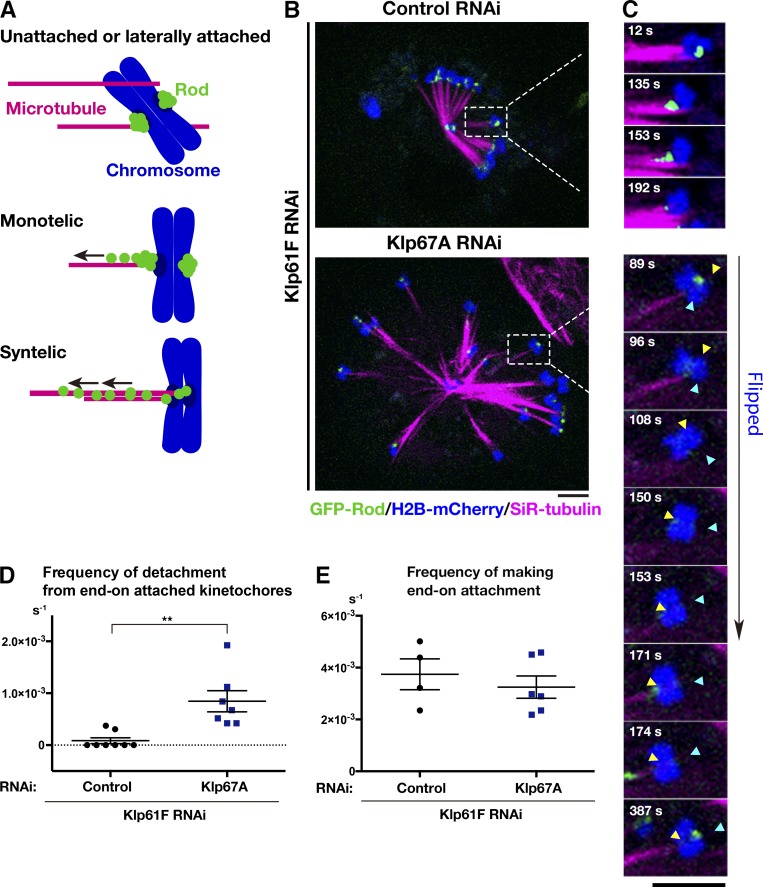
Figure 4.
Kinesin-8Klp67A is required for stable kinetochore–MT attachment: monopolar spindle. (A) Diagram of GFP-Rod and MT attachment mode. (B) Monopolar spindles formed after RNAi of kinesin-5Klp61F (top) or double kinesin-5Klp61F/kinesin-8Klp67A (bottom). Blue, chromosome (H2B-mCherry); green, GFP-Rod; and magenta, MT (SiR-tubulin). (C) Monotelic-to-syntelic conversion in the control monopolar spindle (top); upon new MT association, GFP-Rod stream was observed along the kinetochore MT (153 s), and subsequently, the GFP signal diminished at the kinetochore (192 s). Chromosome flipping (bottom) was observed in the absence of kinesin-8Klp67A (89–171 s; sister kinetochores are indicated by yellow and blue arrows). Monotelic-to-syntelic conversion was also observed for this chromosome (96 s). (D) Increased frequency (event no. per chromosome per second) of MT detachment in the absence of kinesin-8Klp67A (**, P < 0.009; Welch’s t test). Events were counted when MTs terminated end-on attachment. RNAi and imaging were performed four times, data were quantitatively analyzed twice, and the two datasets were combined. Each dot in the graph represents mean frequency for a cell. A total of 86 chromosomes in eight cells (control) and 56 chromosomes in eight cells (kinesin-8Klp67A RNAi) were analyzed. (E) Frequency (event no. per chromosome per second) of newly acquired end-on attachment that was indicated by GFP-Rod stream along kinetochore MTs. The attachment number was divided by the total time the chromosomes spent in a monotelic or unattached state. Each dot in the graph represents mean frequency for a cell. A total of 18 chromosomes in four cells (control) and 18 chromosomes in six cells (kinesin-8Klp67A RNAi) were analyzed. Error bars indicate SEM. Bars, 5 µm.