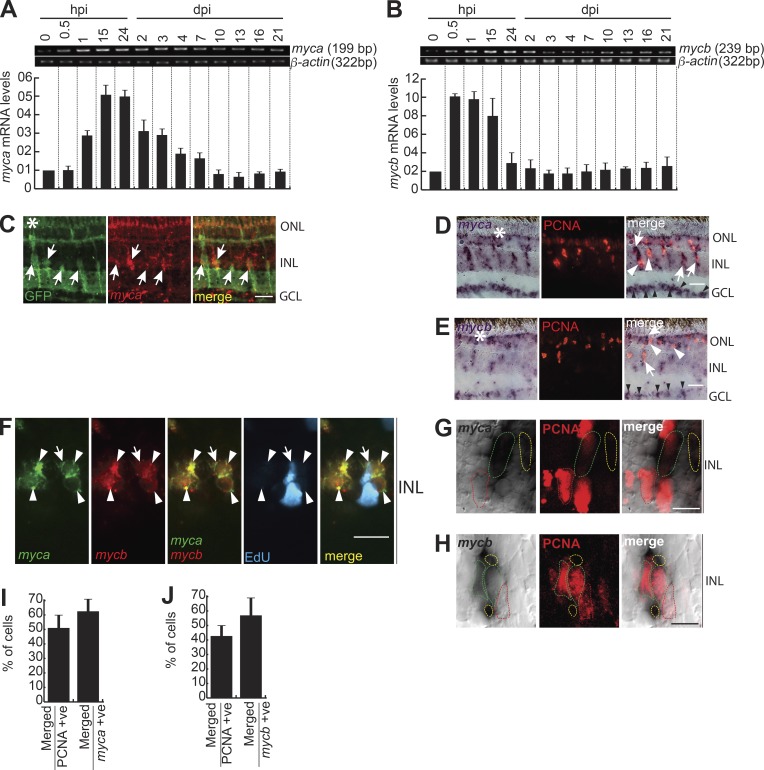
Figure 1.
Myc genes are rapidly induced in the injured retina. (A and B) RT-PCR (top) and qPCR (bottom) were used to assay injury-dependent myca (A), and mycb (B) gene expressions; n = 6 biological replicates. (C) FISH and IF microscopy show expression of myca mRNA and GFP in retina of 1016 tuba1a:gfp transgenic fish at 4 dpi. (D and E) ISH and IF microscopy show that myca (D) and mycb (E) mRNA is expressed in PCNA+ MGPCs and neighboring cells at 4 dpi. The white arrows indicate colabeled cells in D and E, and white arrowheads in D and E identify myca− and mycb− but PCNA+ cells near injury site. Black arrowheads indicate GCL-specific myca and mycb expression. (F) FISH microscopy shows coexpression of myca and mycb mRNA in BrdU+ cells and vicinity. The white arrow indicates EdU+, myca+, and mycb+ cells, and arrowheads mark myca+ and mycb+ cells. (G–J) A single 0.5-µm-thick Z section shows myca (G) and mycb (H) in 4-dpi retina; dotted outline in red shows PCNA+ myca−/mycb− cells, green shows colabel with PCNA and myca/mycb, and yellow indicates myca+/mycb+ but PCNA− cells; and the percentage colabeling with PCNA is quantified (I and J); n = 5 biological replicates. Bars, 10 µm; white asterisks mark the injury sites. ONL, outer nuclear layer; INL, inner nuclear layer (C–H).