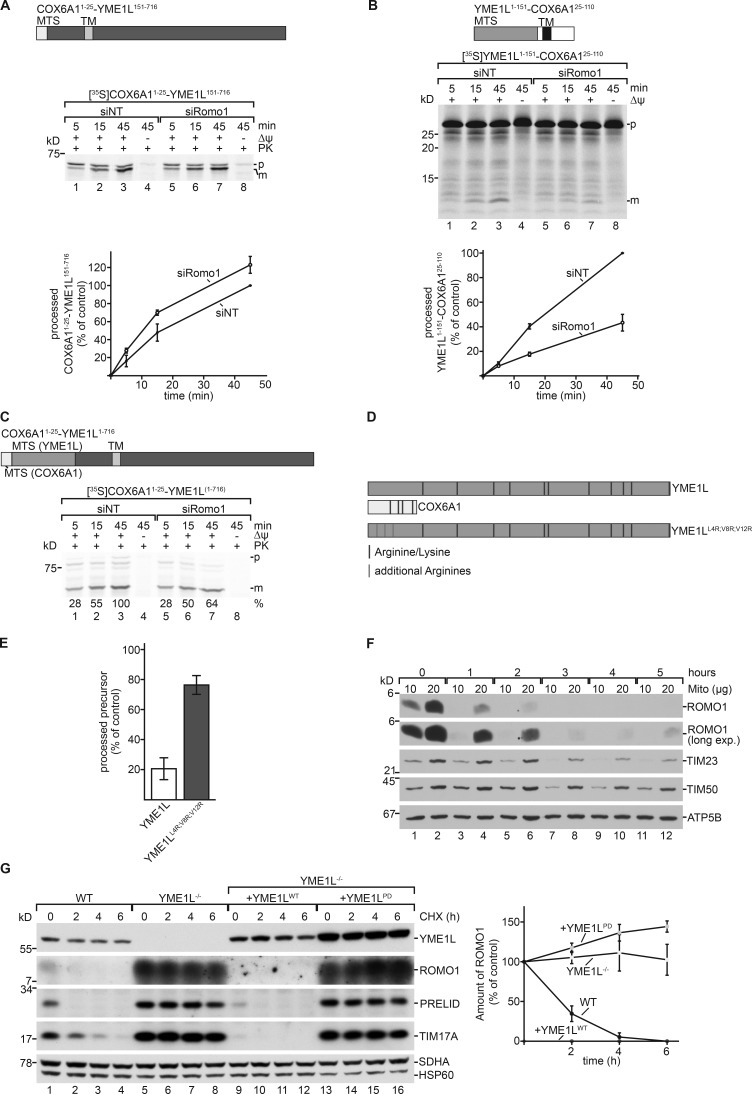
Figure 6.
The presequence of YME1L renders its import dependent on ROMO1. (A and B) The MTS of COX6A1 (first 25 amino acids) was fused to the mature part of YME1L (151 to end; A). The MTS of YME1L (first 151 amino acids) was fused to the mature part of COX6A1 (25 to end; B). Indicated 35S-labeled precursors were imported into isolated energized mitochondria and analyzed by SDS-PAGE and autoradiography. Import of siNT sample at the longest time point was set to 100% (means ± SEM; n = 3) is shown. p, precursor; m, mature protein. (C) The MTS of COX6A1 (first 25 amino acids) was fused to the precursor form of YME1L (1–716). 35S-labeled precursor proteins were imported into isolated energized mitochondria, treated with PK, and analyzed by SDS-PAGE and autoradiography. Import of siNT sample at the longest time point was set to 100%. p, precursor; m, mature protein. (D) Schematic representation of arginines and lysines in presequences of YME1L and COX6A1. Schematic depiction of precursor used in E. Three nonpolar amino acids were replaced by arginines as indicated. (E) 35S-labeled YME1L (Fig. 5 E) and 35S-labeled YME1LL4R;V8R;V12R were imported into isolated energized mitochondria and analyzed by SDS-PAGE and autoradiography. Import of respective siNT sample at 15 min was set to 100% (mean ± SEM; n = 3). (F) HEK293T WT cells were treated with emetine for indicated time points, lysed, and analyzed by SDS-PAGE and immunoblotting. (G) HEK293 WT, YME1L−/−, and YME1L−/− cells expressing WT or proteolytic-dead YME1L in the presence of tetracycline were treated with cycloheximide (CHX; 100 µg/ml) for the indicated time points. YME1L-dependent degradation of substrates was monitored by SDS-PAGE and immunoblotting. The mean ROMO1 protein band intensity was quantified ± SEM (n = 3).