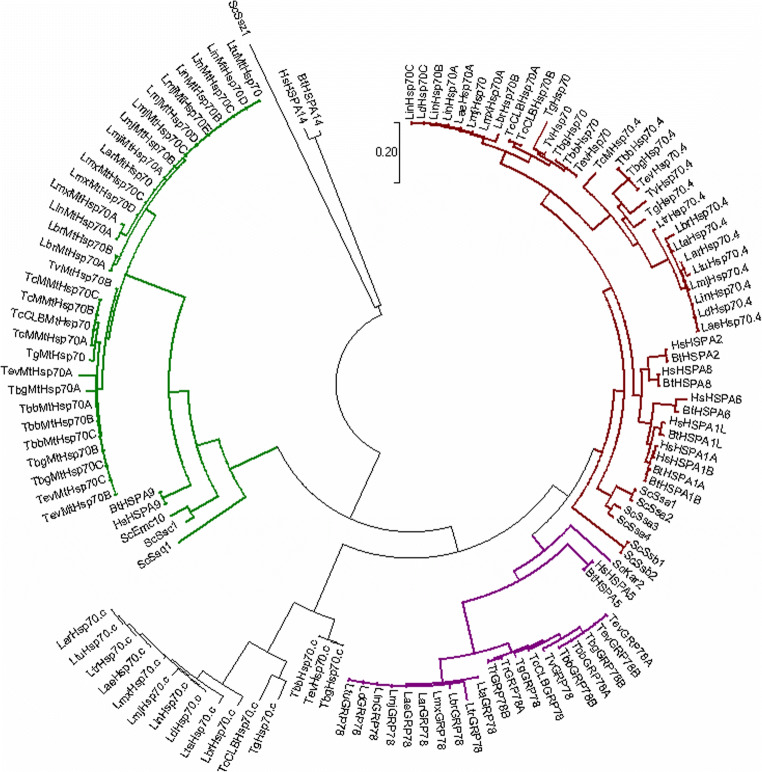
Fig. 1.
Phylogenetic analysis of the Hsp70 superfamily from T. brucei in relation to human and selected kinetoplastid parasites. Multiple-sequence alignment of the full-length amino acid sequences of the Hsp70/HSPA gene families in human and selected kinetoplastid parasites. The multiple sequence alignment provided in Fig. S1 was performed using the in-built ClustalW program (Larkin et al. 2007) with default parameters on the MEGA 7 software (Kumar et al. 2016). The phylogenetic tree was constructed by MEGA 7 using the Maximum-likelihood method based on the Jones–Taylor–Thornton (JTT) matrix-based model of amino acid substitution (Jones et al. 1992) with gamma distribution shape parameter (G). The alignment gaps were excluded from the analysis, and the number of amino acid sites used to construct the tree numbered 363. Bootstrap analysis was computed with 1000 replicates. Accession numbers for the T. b. brucei (Tbb), T. b. gambiense (Tbg), T. cruzi (TcCLB; CL Brener Esmeraldo), C. fasciculata (Cf), B. saltans (Bs), and L. major (Lmj) Hsp70 and Hsp110 sequences can be found in Table 1. Accession numbers for human (Hs; H. sapiens) and other kinetoplastid HSPA/Hsp70 and HSPH/Hsp110 sequences are provided in Table S1. The subcellular localisation for Hsp70s is indicated by coloured branches. Brown: cytosolic; purple: endoplasmic reticulum; and green: mitochondrion. Scale bar represents 0.2 amino acid substitutions per site (colour figure online)