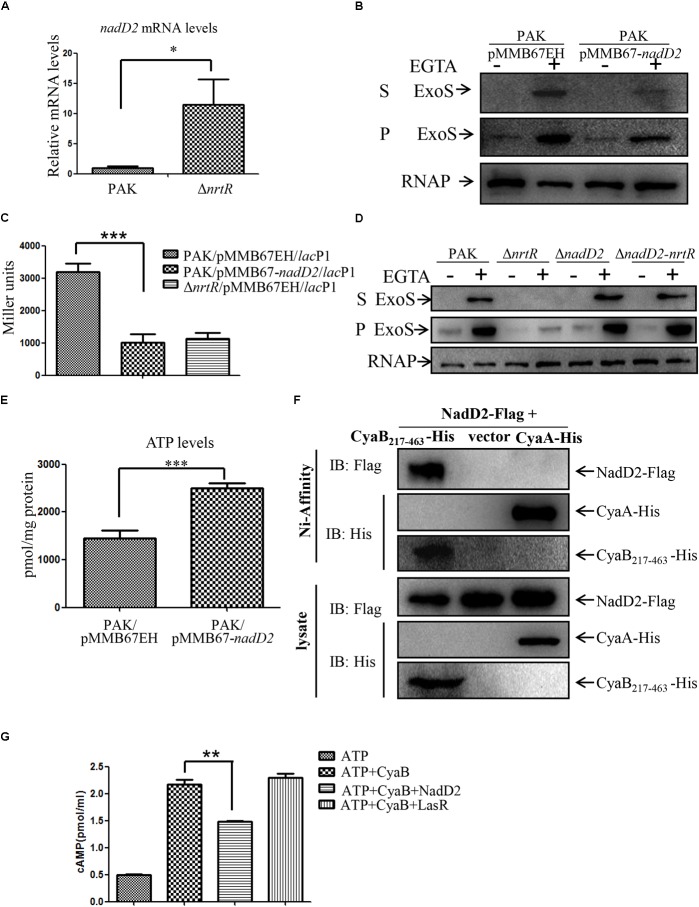
FIGURE 6.
Increased expression of NadD2 contributes to the decreased intracellular cAMP and expression of T3SS in the ΔnrtR mutant. (A) Relative mRNA levels of nadD2. Total RNA of indicated strains was isolated and mRNA levels of nadD2 were determined by real time PCR with rpsL serving as an internal control. Data represents the mean ± standard deviation. ∗p < 0.05 by Student’s t-test. (B) PAK containing nadD2 driven by a tac promoter or the empty vector pMMB67EH were grown to an OD600 of 1.0 in LB containing 1 mM IPTG with or without 5 mM EGTA. Proteins from equivalent bacterial cells of indicated strains were separated by SDS-PAGE and probed with an antibody against ExoS or RNA polymerase beta subunit. S, supernatant; P, pellet. (C) The cAMP levels were decreased in the PAK/pMMB67-nadD2 strain. β-Galactosidase assay was used to examine the LacZ level driven by the lacP1 promoter in indicated strains with 1 mM IPTG induction. Error bars represent standard deviations. ∗∗∗P < 0.001 by Student’s t-test. (D) Indicated bacteria were cultured to an OD600 of 1.0 in LB with or without 5 mM EGTA. Proteins in supernatants and pellets from equivalent bacterial cells were loaded onto SDS-PAGE gels and probed with an antibody against ExoS or RNA polymerase beta subunit. S, supernatant; P, pellet. (E) ATP levels in indicated strains. ∗∗∗P < 0.001 by Student’s t-test. (F) Interaction between NadD2 and CyaB217-463 or CyaA. ΔnadD2 carrying pUCP24-nadD2-Flag with pMMB67EH-cyaB217-463-His, pMMB67EH-cyaA-His or pMMB67EH were grown to an OD600 of 0.6 and incubated with 1 mM IPTG for 18 h at 16°C. Bacteria were lysed and subjected to chromatography with Ni-NTA beads. His-tagged CyaB217–463 or CyaA and FLAG-tagged NadD2 were detected by Western blot assay. (G) Inhibition of CyaB adenylyl cyclase activity by NadD2. 2.0–2.25 μg CyaB was incubated with 5 mM ATP. 4.5–5.5 μg NadD2 or LasR was added as indicated. After 30 min at 30°C, the cAMP levels were measured using an ELISA kit. Error bars represent standard deviations. ∗∗P < 0.01 by Student’s t-test.