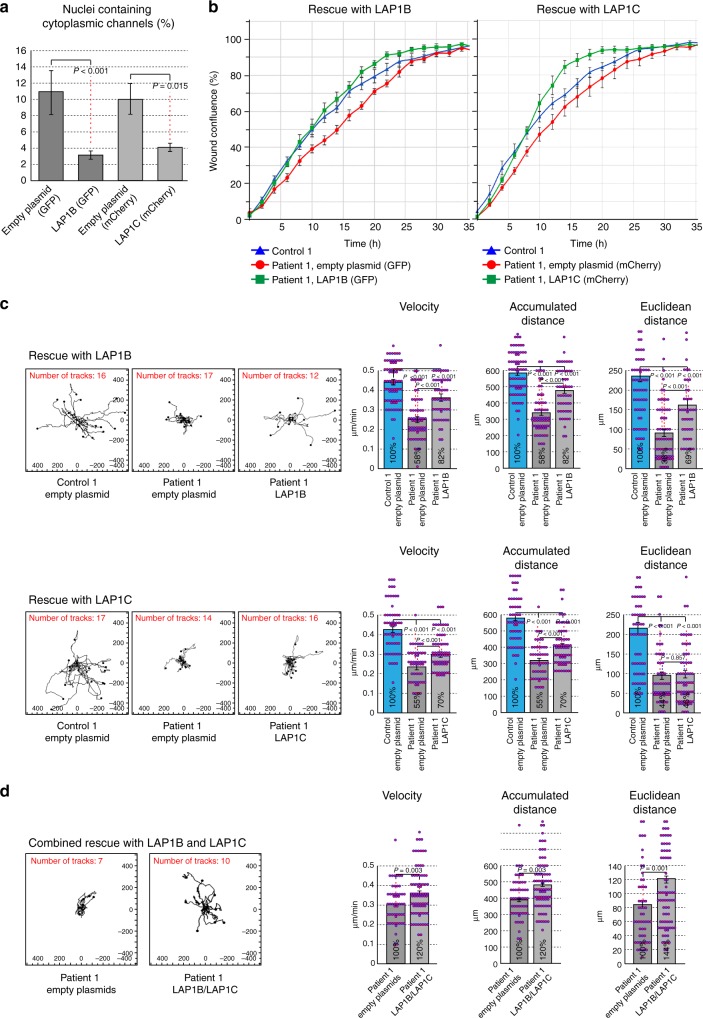
Fig. 7.
Rescue of nuclear morphology and cell motility defects by transduction with lamina-associated polypeptide 1B (LAP1B) and LAP1C coding constructs. Multicistronic lentiviral vectors encoding LAP1B or LAP1C driven by the EF1a promoter and containing two internal ribosome entry site sequences, EGFP or mCherry, and a puromycin resistance marker were constructed and lentiviral particles were prepared as described in Methods. Control or Patient 1 primary fibroblast lines were transduced with lentiviral particles containing LAP1B-coding, LAP1C-coding, or empty plasmid controls. Following several rounds of antibiotic selection and visual verification of fluorescent marker expression in all cells, the stably transfected cell lines were analyzed in three separate assays all demonstrating partial but significant reversal of the defects. a The occurrence of cytoplasmic channels in the nuclei of stably transfected Patient 1 cell lines was analyzed by widefield immunofluorescence, as in Fig. 3b (n = 110 in 2 independent experiments, performed in duplicates; bars indicate SEM). b Wound closing activity was analyzed simultaneously for three cell lines at a time in the scratch wound assay, as in Fig. 6a. Original untransfected control fibroblasts were compared to Patient 1 cell lines stably transfected with the LAP1B-coding or LAP1C-coding and the equivalent empty plasmid controls. Wound confluence was graphed over time with images recorded every 2 h for a total duration of 72 h, showing the means ± SEM for six replicates in each cell line. c Random two-dimensional (2D) cell motility was analyzed by trajectory plots and quantitative analysis, as in Fig. 6b. n = 73 (Control, empty plasmid), n = 62 (Patient 1, empty plasmid), n = 52 (Patient 1, LAP1B); n = 58 (Control, empty plasmid), n = 51 (Patient 1, empty plasmid), n = 67 (Patient 1, LAP1C); bars indicate SEM. d An attempt to achieve combined rescue with both LAP1B and LAP1C coding constructs. A direct comparison is shown between Patient 1 cells stably transfected with the two empty plasmids and with the two LAP1-coding plasmids. Note that this experiment cannot be conducted under optimal conditions because of the lack of a second selection marker, although GFP and mCherry were both observed in all the cells analyzed by trajectory plots. Random cell motility in 2D was analyzed as above. Although rescued motility is observed, the quantitative analysis parameters do not show a synergistic effect beyond what is achieved by the LAP1B construct alone (c, top row). n = 53 (Patient 1, empty plasmids), n = 78 (Patient 1, LAP1B+LAP1C); bars indicate SEM