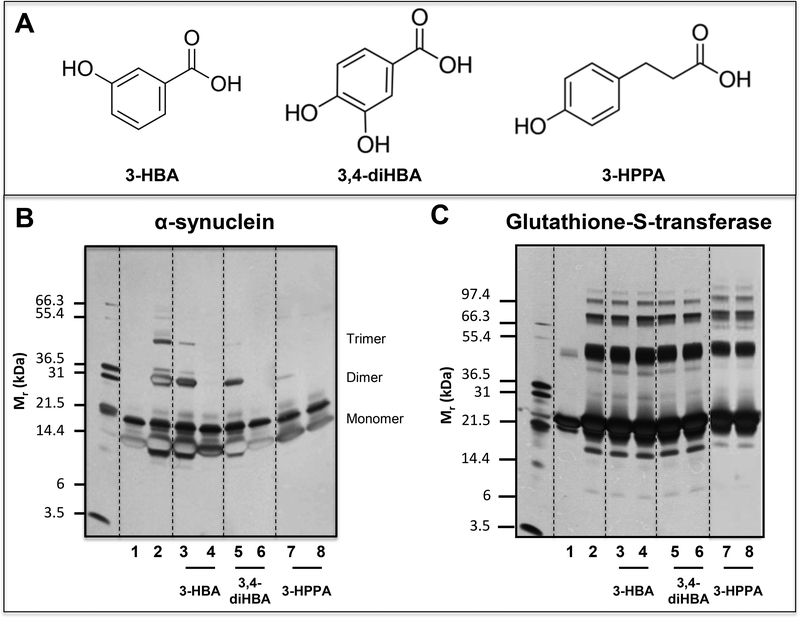
Fig. 3. Select brain accumulating, FRP-derived microbial phenolic acid metabolites potently interfere with protein-protein interactions among α-synuclein peptides.
Using the PICUP protocol, we assess effects of three structurally closely related phenolic acid metabolites, 3-HBA, 3,4-diHBA, and 3-HPPA on the initial protein-protein interactions necessary for the assembly of monomeric α-synuclein into multimeric forms. PICUP assays were conducted in the presence of vehicle or individual phenolic acids at a low dose (LD, equal molar concentration of phenolic acid relative to α-synuclein) or at a high dose (HD, 4-fold higher molar concentration of phenolic acid relative to α-synuclein). Monomeric and cross-linked oligomeric α-synuclein forms were resolved by SDS-PAGE and visualized by silver staining. We confirmed that the addition of phenolic acids did not lead to observable change in the pH of the reaction mixture. (A) Molecular structures of the 3-HBA, 3,4-diHBA, and 3,4-diHBA. (B) Effects of individual phenolic acids on the oligomerization of monomeric α-synuclein peptides. (C) Control PICUP studies using glutathione-S-transferase confirming that the addition of 3-HBA, 3,4-diHBA or 3-HPPA at both and LD or the HD did not interfere with the PICUP cross-linking mechanism. In (B, C) Lane 1, peptide (α-synuclein or glutathionne-S-transferase) without cross linking; lane 2, peptide cross-linked in the presence of vehicle; lane 3, peptide cross-linked in the presence of a LD of 3-HBA; lane 4, peptide cross-linked in the presence of a HD of 3-HBA; lane 5, peptide cross-linked in the presence of a low dose of 3,4-diHBA; lane 6, peptide cross-linked in the presence of a HD of 3,4-diHBA; lane 7, peptide cross-linked in the presence of a LD of 3-HPPA; lane 8, peptide cross-linked in the presence of a HD of 3-HPPA. Each of the gel shown is representative of three independent experiments.