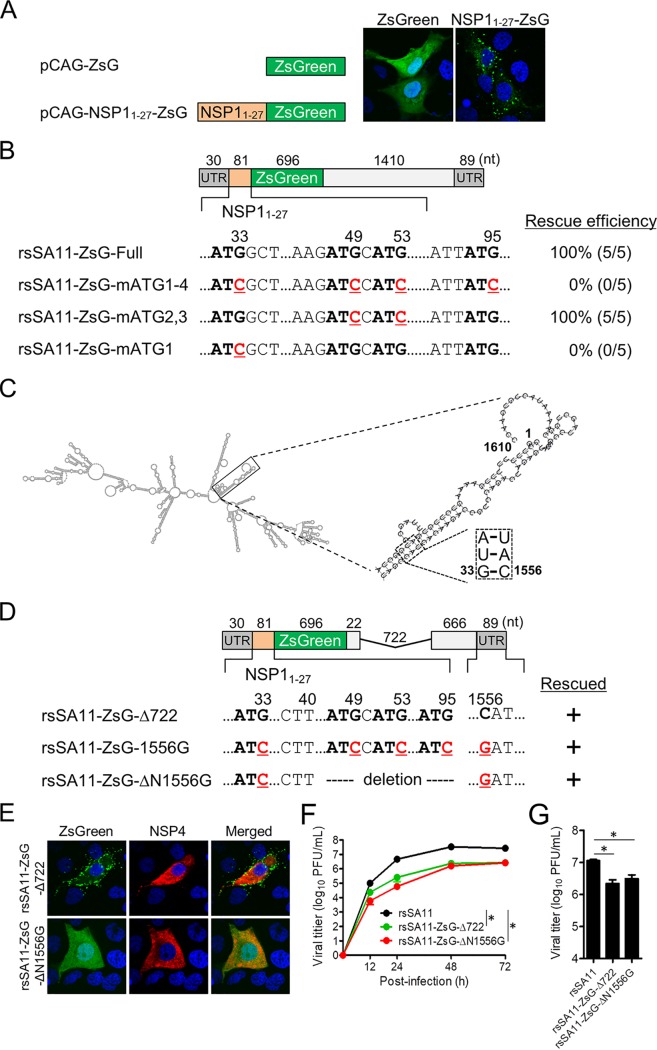
FIG 7.
Modification of the reporter gene. (A) Cellular localization of a recombinant ZsG protein expressed with/without the NSP11–27 peptide plasmid vector. (B) Partial nucleotide sequence of the NSP11–27-ZsG gene used for reverse genetics. Four ATG triplets (bold) and ATG-to-ATC substitutions (red, underlined) in the NSP11–27 coding sequence are shown. The results of virus rescue and the rescue efficiencies of recombinant viruses harboring each reporter gene are shown. Data represent the mean values of five independent rescue experiments. (C) Predicted secondary structure of wild-type NSP1 mRNA. Double-strands formed by the 5′- and 3′-terminal regions are shown in the insets. Base paring at positions 33 and 1556 within the stem structure are shown in the square. Nucleotide positions are numbered according to the wild-type NSP1 gene sequence. (D) Schematic showing the NSP1-ZsG-1556G and NSP1-ZsG-ΔN1556G constructs. Nucleotide positions are numbered according to the wild-type NSP1 sequence. (E) MA104 cells were infected with rsSA11-ZsG-Δ722 or rsSA11-ZsG-ΔN1556G. Viral NSP4 was detected using rabbit anti-NSP4 and an anti-rabbit-IgG-CF594 conjugate. Nuclei were stained with 4′,6-diamidino-2-phenylindole. (F and G) Replication kinetics of rsSA11-ZsG-ΔN1556G. MA104 cells were infected with rsSA11, rsSA11-NLuc-Δ722, or rsSA11-ZsG-ΔN1556G virus at MOI of 0.001 PFU per cell and incubated for various times (F) or at an MOI of 5 PFU per cell and incubated for 16 h (G). Infectious virus titers in cell lysates were determined by the plaque assay. Data are expressed as the means ± standard deviations (n = 3). Statistical significance was determined by one-way ANOVA and Tukey’s multiple-comparison posttest. *, P < 0.05.