FIG 1.
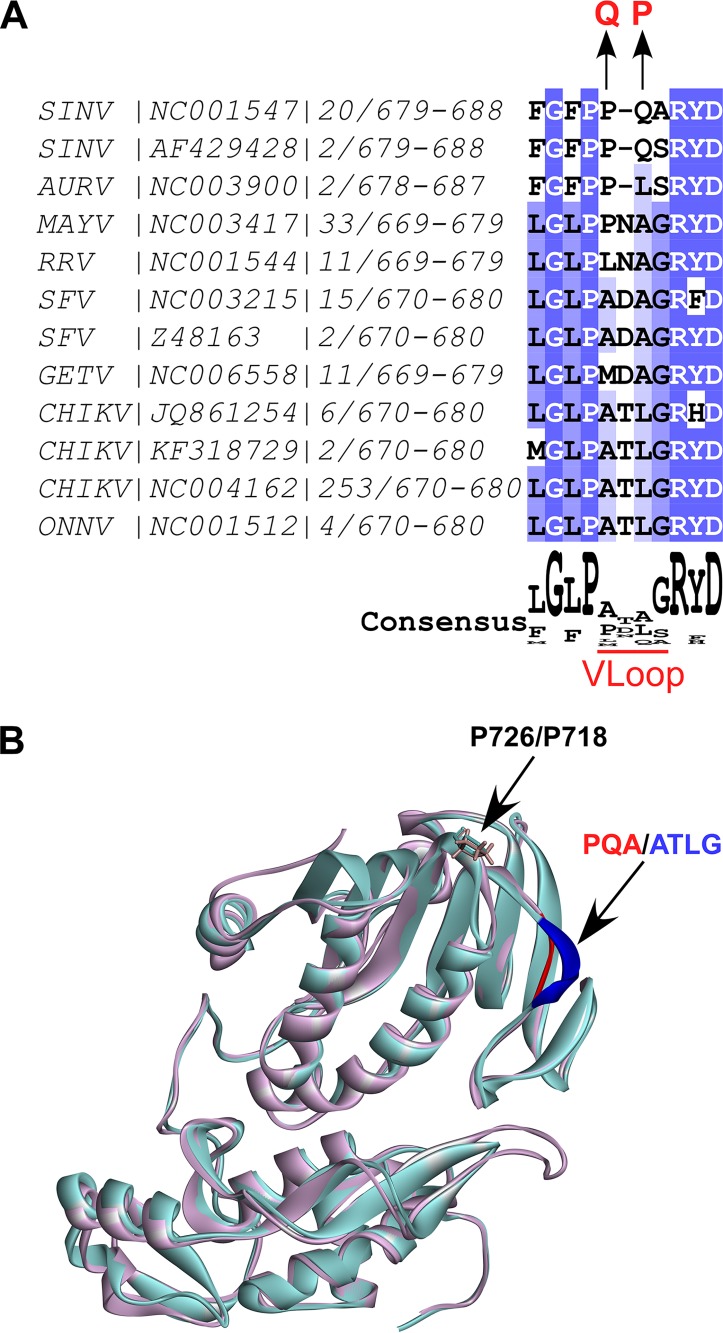
CHIKV nsP2-specific VLoop demonstrates high variability and is located on the surface of the carboxy-terminal SAM MTase-like domain. (A) The nsP2 protein sequences of 365 OW alphaviruses were downloaded from Virus Pathogen Resources (ViPR). The sequences were aligned using Muscle in Jalview. The fragments of nsP2 corresponding to aa 679 to 688 of SINV were aligned, and then the redundant sequences and those present in a single strain were deleted. The GenBank accession number for the representative strain, numbers of the strains included into alignment for each group, and amino acid positions in nsP2 are indicated. Mutations identified in attenuated SINV are shown above the alignment. (B) The 3D structures of CHIKV (PDB code 3TRK) and SINV (modeled based on PDB code 4GUA) were superimposed in Discovery Studio Visualizer using sequence alignment. The structure of CHIKV nsP2 is presented as a light magenta solid ribbon. The structure of SINV nsP2 is presented as a light turquoise solid ribbon. P726 (SINV) and corresponding P718 (CHIKV) are presented as sticks. CHIKV-specific VLoop is colored in blue, and SINV-specific VLoop is colored in red. Abbreviations: AURV, Aura virus; CHIKV, chikungunya virus; GETV, getah virus; MAYV, Mayaro virus; RRV, Ross River virus; ONNV, o’nyong nyong virus; SFV, Semliki Forest virus; SINV, Sindbis virus.