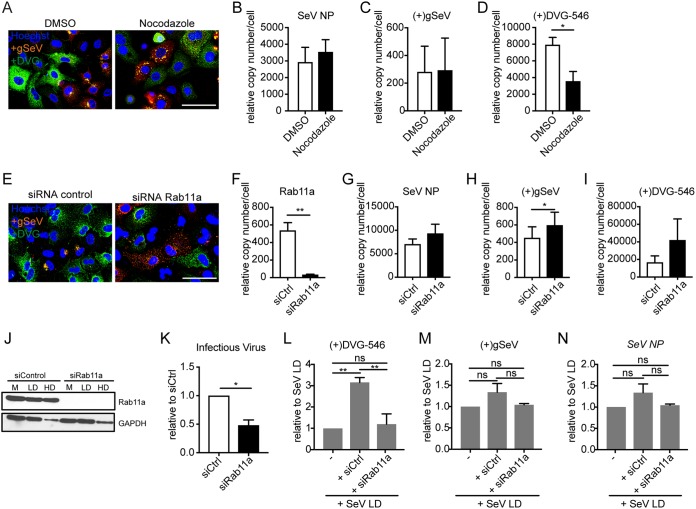
FIG 6.
Cytoplasmic distribution of DVGs in DVG-high cells is independent of microtubule integrity and Rab11a expression. (A) A549 cells infected with SeV-HD and treated with nocodazole at 4 h postinfection. Positive-sense viral RNA-FISH was performed at 24 h postinfection. Wide-field images were captured with a 63× objective. Deconvolved, extended focus is shown. Bar = 100 μm. Images are representative of results from four independent experiments. DMSO, dimethyl sulfoxide. (B to D) RT-qPCR for SeV NP (B), (+)gSeV (C), and (+)DVG-546 (D) transcripts relative to GAPDH at 24 h postinfection. Data are represented as means ± SEM of results from three independent experiments. *, P < 0.05 by a paired t test. (C) A549 cells transfected with siRNA targeting Rab11a or scrambled control siRNA (siCtrl) prior to infection, infected with SeV-HD for 24 h, and then subjected to positive-sense viral RNA-FISH. Wide-field images were captured with a 63× objective. Deconvolved, extended focus is shown. Bar = 100 μm. Images are representative of results from four independent experiments. (F to I) RT-qPCR for Rab11a (F), SeV NP (G), (+)gSeV (H), and (+)DVG-546 (I) transcripts relative to GAPDH at 24 h postinfection. Data are represented as means ± SEM of results from seven independent experiments. *, P < 0.05; **, P < 0.005 by a paired t test. (J) Western blotting for Rab11a protein with a GAPDH loading control in mock (M), SeV-LD (LD), and SeV-HD (HD) infections. (K) Relative levels of infectious virus by TCID50 normalized to an siRNA control. Data are represented as means ± SEM of results from three independent experiments. *, P < 0.05 by a paired t test. The supernatant from control siRNA and Rab11a siRNA KD (knockdown) cells was adjusted to an MOI of 1 TCID50/cell with SeV-LD, and LLCMK2 cells were infected for 24 h. (L to N) RT-qPCR for SeV Cantell-specific DVG-546 (L), (+)gSeV (M), and SeV NP (N) mRNAs relative to GAPDH. Data are represented as means ± SEM of results from three independent experiments normalized to values for SeV-LD alone. **, P <0.005 by one-way ANOVA followed by a Bonferroni post hoc test.