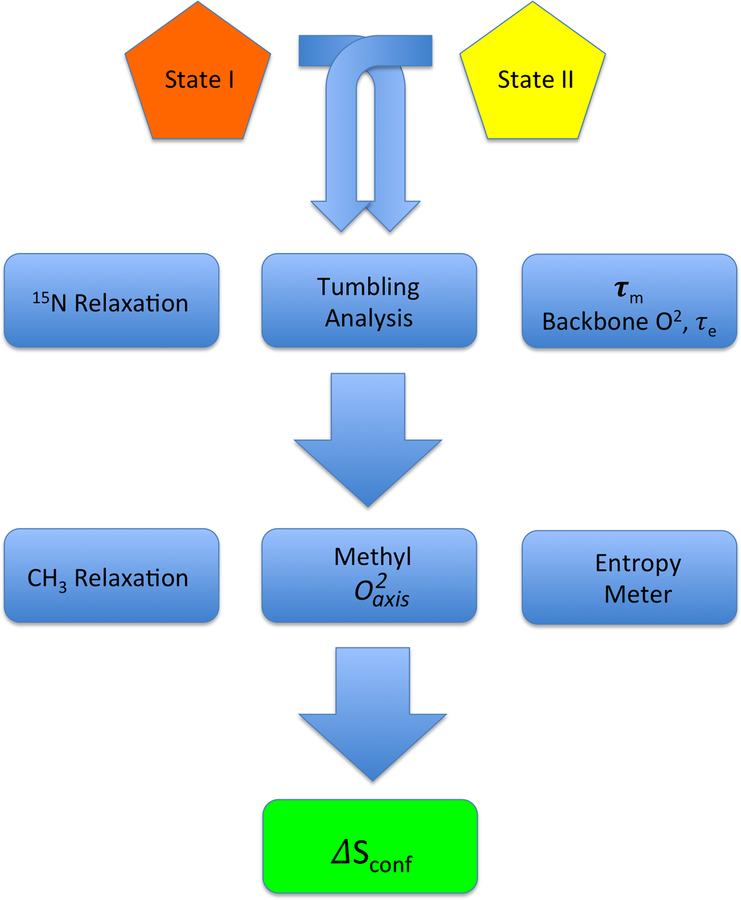
Figure 1.
Simplified flow chart illustrating the key steps in determining protein conformational entropy from NMR relaxation measurements. Macromolecular tumbling is best characterized using relatively rigid components of the protein i.e. the backbone. Model-free analysis provides information about the dynamical character the backbone in addition to the appropriate tumbling model and parameters to employ subsequently. Methyl-relaxation is generally sufficient to carry through to an analysis of conformational entropy. Determination of absolute entropy is fraught with difficult (see text) and only differences in conformational entropy should determined, hence the requirement to compare two states (e.g. protein with and without ligand, etc.).