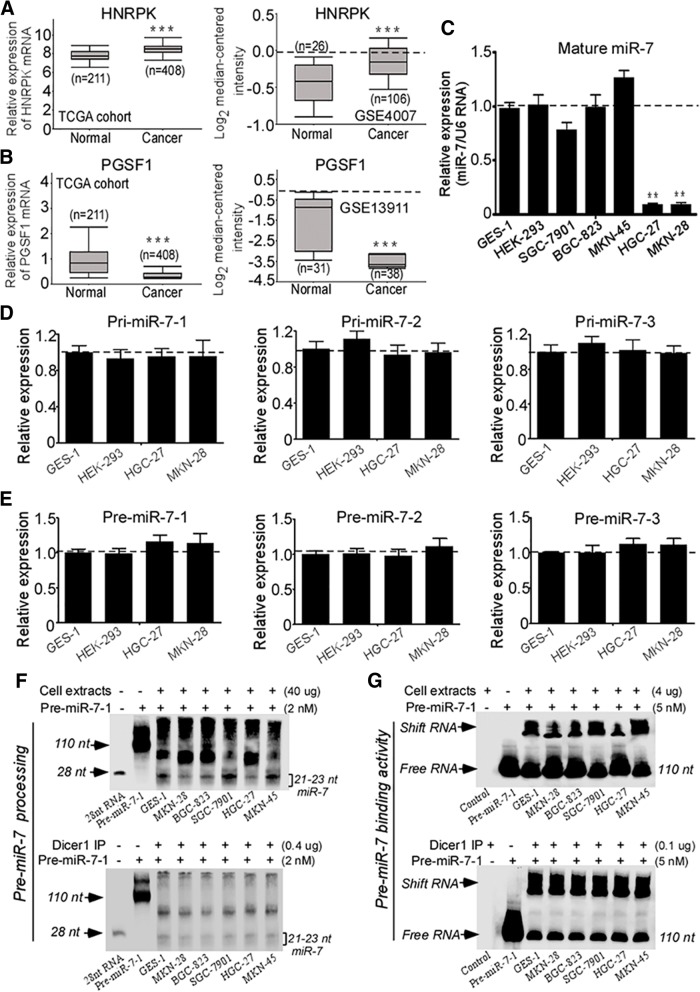
Fig. 3.
Impaired pre-miR-7 processing is responsible for downregulated miR-7 in GC cells. a Upregulated HNRPK (miR-7-1 host gene) mRNA expression in TCGA STAD (n = 619, P < 0.001) and in the NCBI/GEO/GSE4007 database l (n = 132, P < 0.001). b Downregulated PGSF1 (miR-7-3 host gene) mRNA expression of GC in TCGA STAD (n = 619, P < 0.001) and in the NCBI/GEO/GSE13911 database (n = 69, P < 0.001). c Expression of mature miR-7 in five GC cell lines relative to GES-1 and HEK-293 cells. Total RNA was extracted from indicated cells. Expression of mature miR-7 was detected by real-time PCR. U6 RNA was used as an internal control. d-e Expression of pri-miR-7 and pre-miR-7 isoforms in GC cells. Pri-miR-7 isoforms (pri-miR-7-1,2,3) (d) and pre-miR-7 isoforms (pre-miR-7-1,2,3) (e) were quantitated by modified real-time PCR in HGC-27 and MKN-28 cells compared with GES-1 and HEK-293 cells. Human 18S rRNA was used as an internal control. f Pre-miR-7-1 processing assay. Pre-miR-7-1 RNA (2 nM) was incubated in processing buffer with cell extracts (Uper panel) or Dicer1 containing IP complex (Lower panel) respectively. The 110 nt pre-miR-7-1 and mature ~ 21-23 nt miR-7 were detected by RNA gel blot after processing action. g Pre-miR-7-1 binding assay. Pre-miR-7-1 RNA (5 nM) was incubated in binding buffer with cell extracts (Uper panel) or Dicer1 containing IP complex (Lower panel) respectively. RNA gel blot was used to detect shifted binding pre-miR-7-1 RNA and free pre-miR-7-1 RNA (110 nt) after binding action. Data are presented as mean ± SD of three independent experiments. * P < 0.05; ** P < 0.01 between the indicated two groups determined by paired student’s t test