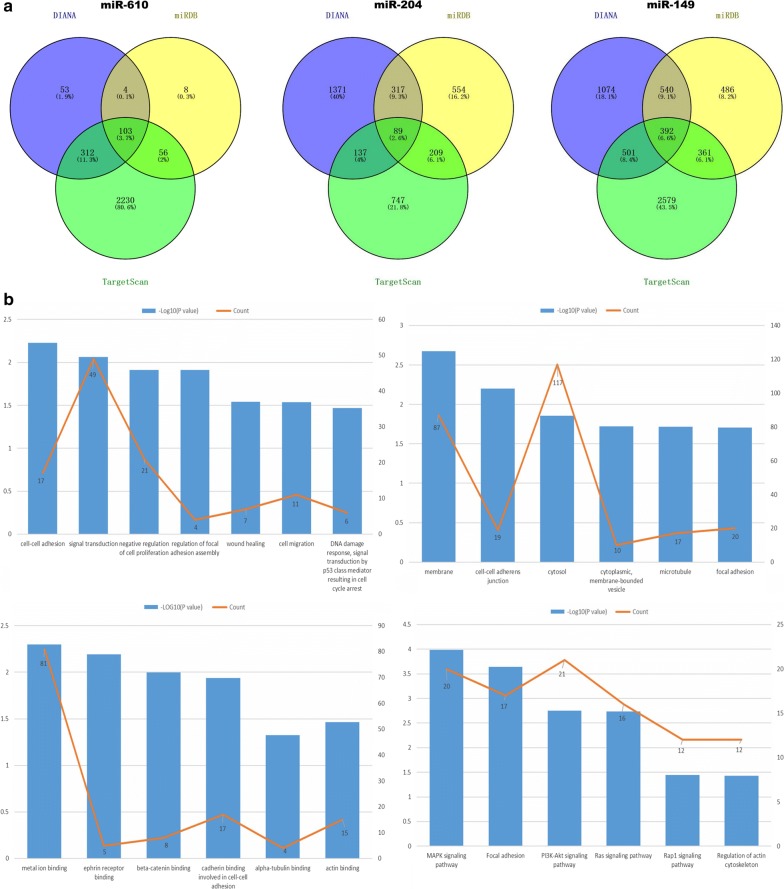
Fig. 1.
Target gene prediction and function analysis. a The common genes targeted by the three miRNAs as predicted by DIANA, miRDB, and TargetScan. b Biological process analysis (upper left) indicated that the genes are associated with cell–cell adhesion, signal transduction, negative regulation of cell proliferation, regulation of focal adhesion assembly, wound healing, and cell migration. Cellular component analysis (upper right) showed that the genes are associated with cell–cell adherent junctions, membranes, focal adhesion, cytosol, and microtubules. Molecular function analysis (lower left) indicated that the genes are involved in metal ion binding, cadherin binding cell–cell adhesion, and beta-catenin binding. Biological pathway analysis (lower right) revealed that the genes participate in PI3K-Akt, MAPK, and Ras signaling pathways