Figure 1.
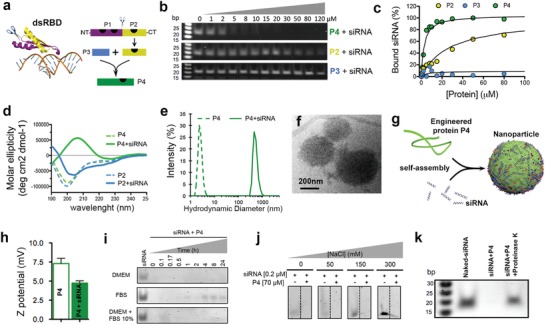
Design of peptide with amplified siRNA binding via self‐assembly. a) Scheme showing crystal structure of a dsRNA‐binding domain from X. laevis (see ref. 15) (left). C‐terminus RNA binding motif P2 (in yellow) was extracted from the dsRBD and fused to a self‐assembly enhancer peptide P3 to obtain P4, a peptide with amplified siRNA binding (right). Black marks on the peptide schematics are dsRNA‐binding sites according to the crystal structure. b) Electrophoresis gels of peptides bound to siRNA. c) Plot of percentage of peptide‐bound siRNA derived from the electrophoresis gels. d) Plot of molar ellipticity (circular dichroism) of P4 and P2 peptides alone (dashed lines) and with siRNA (solid lines). e) Hydrodynamic size of P4 peptide alone (dashed line) and with siRNA (solid line). f) P4–siRNA particles in solution imaged by cryo‐TEM. g) Schematic representation of an unstructured P4 polypeptide that binds siRNA to assemble into particles. h) ζ‐potential of P4 peptide alone (empty bar) and with siRNA (solid bar). i) Electrophoresis gels showing the release and digestion of siRNA bound to peptide P4 when incubated in different media, DMEM (top), FBS 100% (middle), and DMEM + FBS 10% (bottom). j) Electrophoresis gels showing the release of siRNA from siRNA–P4 particles in aqueous solutions as a function of NaCl concentration. k) Electrophoresis gel showing release of siRNA from P4 particles when incubated with proteinase K. Molecular marker (first lane from left to right), naked siRNA (second lane), siRNA–P4 complexes (third lane), and siRNA–P4 complexes incubated with proteinase K for 24 h (fourth lane).
