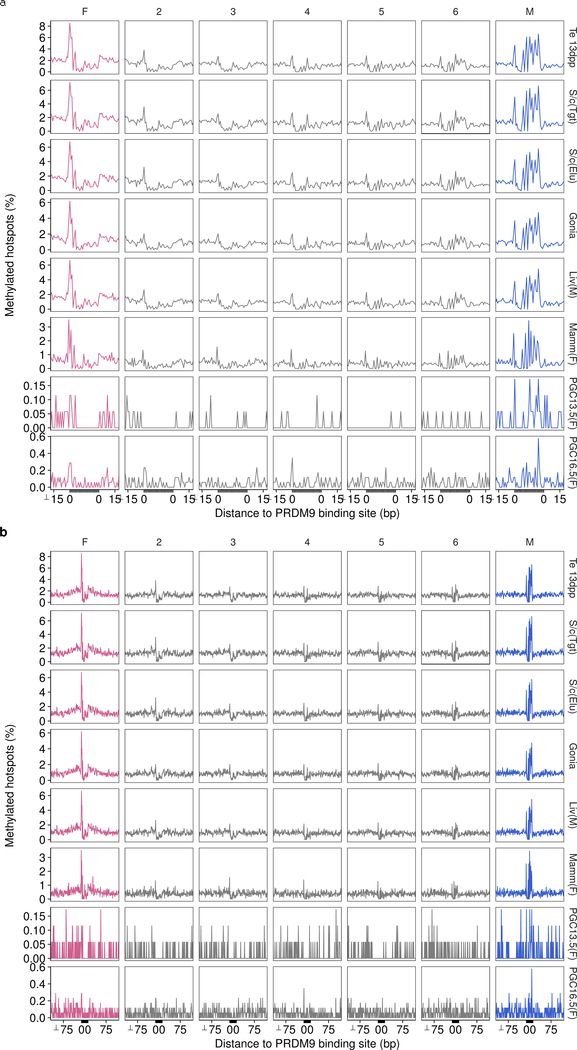
Extended Data Fig 8: DNA methylation at PrBSs is present across tissues and absent in the female germ line.
The pattern of DNA methylation is very similar across cell types and between the sexes. Hotspots are split by the magnitude of sex bias (SSDSO1/SSDST1) into seven sets. Sets are ranked from most female-biased (pink; left) to most male-biased (blue; right) by fold-change. Methylation signal is binarized such that methylation > 0% is considered methylated. Thus, the proportion of all hotspots with methylated cytosine at each position is shown. Variations in the magnitude of the signal may be expected for technical reasons. Plots are anchored by the C57Bl6 PrBS (grey area). Only hotspots with a single PRDM9 binding site are used (see Methods). a, Plot of ± 100 bp to show methylation flanking the PrBS for female-biased hotspots. b, Plot of ± 15 bp to show methylation at the PrBS for male-biased hotspots. Methylation data are from whole genome bisulfite sequencing (WGBS) in tissue derived from whole testis in 13dpp mice40 (Te 13 dpp), from WGBS following H3K4me3 CHiP-Seq in whole adult testis (S/c(Tgt)); Extended Data Fig 1h), from WGBS in elutriated spermatocytes38 (S/c(Elu)), from WGBS in spermatogonia38 (Gonia), from WGBS in tissue from male liver53 (Liv(M)), from WGBS in tissue from female parous basal differentiated mammary gland cells54 (Mamm(F)) and from WGBS in sorted primordial germ cells at 13.5 dpc (PGC13.5(F)) and at 16.5 dpc30 (PGC16.5(F)). WGBS in female PGCs captures the methylation status of the genome in oocytes during meiosis30. 13.5 dpc and 16.5 dpc likely represent earlier and later meiotic prophase I populations, respectively.