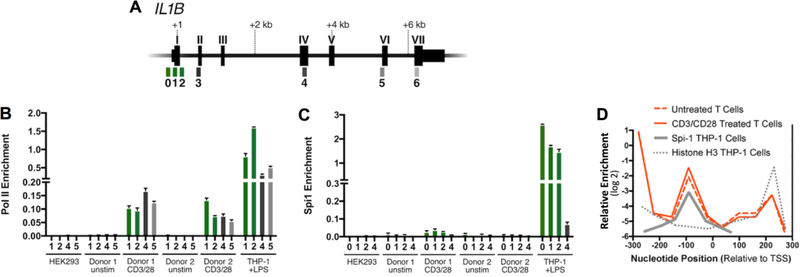
Figure 3. Occupancy of Pol II and Spi1 on the IL1B gene.
(A) Gene schematics with exons labeled as Roman numerals, and qPCR amplicons indicated by Arabic numbers referenced to positions listed in Supplementary Table 1. (B) Pol II and (C) Spi1 ChIP for indicated CD4 T cell populations. For comparison, IL1B-refractory HEK293 cells were used as a negative control, and THP-1 cells stimulated with LPS for 1.5 hours were used as a positive control. CD4 T cells were purified from human lymphoid tissue and activated for 3 days with anti-CD3/CD28 beads and compared to unstimulated CD4 T cells. Promoter-proximal amplicons are shown as green bars, whereas downstream amplicons are shown as gray scale bars. Standard error for each donor from lymphocyte populations represents technical replicates, and control cell lines from cultured biologic replicates. (D) Histone H3 nucleosome ChIP at IL1B promoter in ex vivo CD3/CD28-activated and unstimulated CD4 T cells as described within, are qualitatively compared to the previously reported Spi1 binding site and nucleosome distribution at the IL1B promoter in THP-1 cells[9]. This reveals that a nucleosome is positioned over the Spi1 recognition site in T cells. It has also been reported that Spi1-binding displaces this histone in THP-1 cells, but not in both Spi1-negative 293 cells and the HUT102 T cell line[9]. The qPCR amplicons used are listed in Supplementary Table 1.