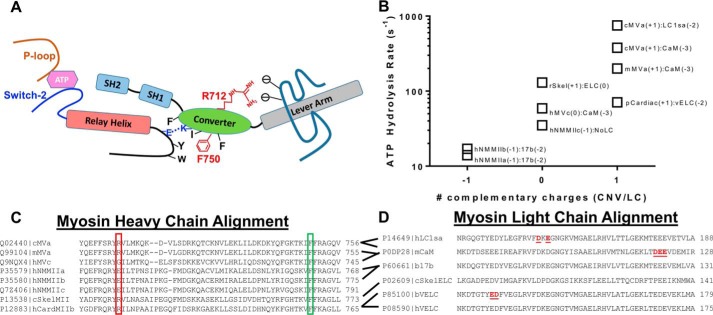
Figure 8.
Summary of allosteric interactions associated with the converter/relay helix/lever arm region. A, a diagram of the key structural elements that allow communication between the nucleotide-binding region and the lever arm (switch II, relay helix/loop, SH1-SH2 helix, converter domain; see “Discussion” for allosteric pathway description). B, graph representing the correlation between the ATP hydrolysis rate constant and potential interactions between the converter domain (Arg-712 site) and the light chain bound to the first IQ motif. C, an alignment of several myosin heavy chains demonstrating conservation of the Arg-712 (red box) and Phe-750 (green box) residues. D, an alignment of several myosin light chains demonstrating negatively charged residues in close proximity to the Arg-712 site. The red underlined residues were found to be in close proximity to the Arg-712 site in several crystal structures (PDB codes 1OE9, 1W7J, 4ZLK and 5N69). The following abbreviations were used for the myosin heavy chains: chicken MYO5A (cMVa), mouse MYO5A (mMVa), human MYO5A (hMVc), human MYH9 (hNMMIIa), human MYH10 (hNMMIIb), human MYH14 (hNMMIIc), chicken MYSS (cSkelMII), rabbit MYSS (rSkel), human MYH7 (hCardMIIb), porcine MYH7 (pCardiac). The following abbreviations were used for the myosin light chains: human MYL6B (hLC1sa), mouse CALM3 (mCaM), bovine ventricular MYL3 (bVELC), human ventricular MYL3 (hVELC), bovine non-muscle MYL6 (bLC17b), chicken skeletal MLRS (cSkelELC). The data in B were taken from the following references: cMVa:LC1sa (30), cMVa:CaM (51), mMVa:CaM (64), rSkel:ELC (65), pCardiac:vELC (66), hMVc:CaM (52), hNMMIIc:NoLC (67), hNMMIIb:17b (68), hNMMIIa:17b (69).