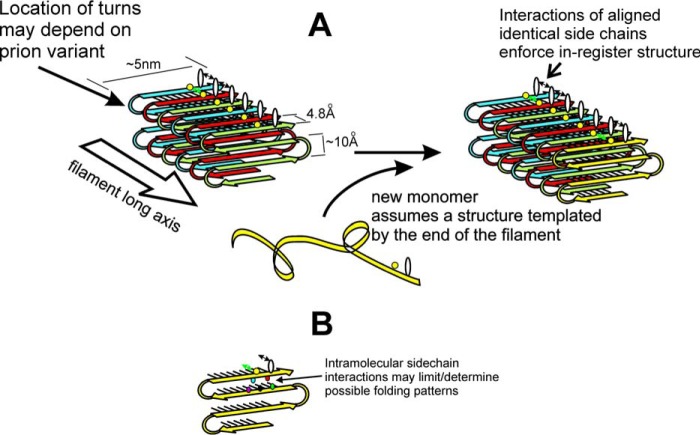
Figure 1.
A, folded parallel in-register β-sheet architecture of yeast prion amyloids Sup35p, Ure2p, and Rnq1p (59–61, 63). It is proposed that prion variants differ largely in the location of the turns in the strands/folds in the β-sheet (B), with favorable H-bonding or hydrophobic interactions among aligned side chains ensuring alignment of identical residues in adjacent molecules. These favorable interactions drive new molecules joining the end of the filament to have their turns in the same places as molecules already in the filament, and this constitutes templating of conformation (68, 69). Reprinted with permission from Ref. 69. This research was originally published in Biochemistry. Wickner, R. B., Edskes, H. K., Bateman, D. A., Kelly, A. C., Gorkovskiy, A., Dayani, Y., and Zhou, A. Amyloids and yeast prion biology. Biochemistry 2013;52:1514–1527. © the American Chemical Society.